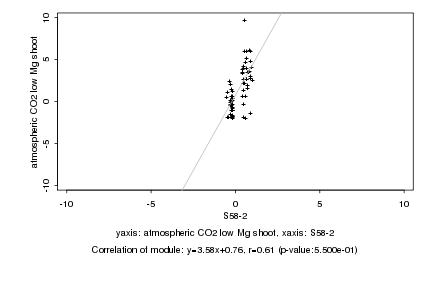
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
S58-2 |
Log2 signal ratio
atmospheric CO2 low Mg shoot |
| 1 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.970 |
2.545 |
| 2 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.908 |
4.084 |
| 3 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.875 |
6.033 |
| 4 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.874 |
4.880 |
| 5 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.870 |
2.847 |
| 6 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.856 |
-1.326 |
| 7 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.845 |
3.079 |
| 8 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.786 |
6.108 |
| 9 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.783 |
3.642 |
| 10 |
252346_at |
AT3G48650
|
[pseudogene] |
0.736 |
3.572 |
| 11 |
264635_at |
AT1G65500
|
unknown |
0.714 |
3.492 |
| 12 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.694 |
2.000 |
| 13 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.656 |
1.583 |
| 14 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.642 |
5.231 |
| 15 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
0.630 |
6.033 |
| 16 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
0.627 |
2.626 |
| 17 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.626 |
2.669 |
| 18 |
260005_at |
AT1G67920
|
unknown |
0.623 |
3.989 |
| 19 |
252309_at |
AT3G49340
|
[Cysteine proteinases superfamily protein] |
0.581 |
-2.021 |
| 20 |
251633_at |
AT3G57460
|
[catalytics] |
0.574 |
4.747 |
| 21 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.553 |
0.683 |
| 22 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.515 |
2.207 |
| 23 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.513 |
9.762 |
| 24 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.493 |
6.013 |
| 25 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.476 |
4.180 |
| 26 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.475 |
1.342 |
| 27 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.466 |
4.049 |
| 28 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
0.455 |
-1.786 |
| 29 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
0.453 |
2.731 |
| 30 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.446 |
2.177 |
| 31 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
0.428 |
-0.306 |
| 32 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.391 |
3.419 |
| 33 |
262542_at |
AT1G34180
|
anac016, NAC domain containing protein 16, NAC016, NAC domain containing protein 16 |
0.389 |
3.550 |
| 34 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.379 |
3.819 |
| 35 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.371 |
0.682 |
| 36 |
262710_at |
AT1G16210
|
unknown |
-0.568 |
0.507 |
| 37 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.516 |
1.113 |
| 38 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.482 |
-1.891 |
| 39 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.425 |
-1.809 |
| 40 |
256021_at |
AT1G58270
|
ZW9 |
-0.380 |
2.398 |
| 41 |
263002_at |
AT1G54200
|
unknown |
-0.340 |
-0.303 |
| 42 |
245792_at |
AT1G32100
|
ATPRR1, PRR1, pinoresinol reductase 1 |
-0.329 |
-0.054 |
| 43 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.328 |
-0.465 |
| 44 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.313 |
2.080 |
| 45 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.303 |
0.179 |
| 46 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
-0.293 |
-1.473 |
| 47 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.292 |
-1.807 |
| 48 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.288 |
-0.430 |
| 49 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.277 |
-0.236 |
| 50 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.276 |
-0.959 |
| 51 |
250281_at |
AT5G13240
|
[transcription regulators] |
-0.276 |
0.631 |
| 52 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
-0.256 |
1.513 |
| 53 |
260241_at |
AT1G63710
|
CYP86A7, cytochrome P450, family 86, subfamily A, polypeptide 7 |
-0.251 |
-0.306 |
| 54 |
245133_at |
AT2G45310
|
GAE4, UDP-D-glucuronate 4-epimerase 4 |
-0.234 |
0.089 |
| 55 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.231 |
0.602 |
| 56 |
263887_at |
AT2G36980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.227 |
-0.662 |
| 57 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.225 |
-1.774 |
| 58 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.225 |
-1.850 |
| 59 |
249865_at |
AT5G22820
|
[ARM repeat superfamily protein] |
-0.220 |
0.372 |
| 60 |
267481_at |
AT2G02780
|
Leucine-rich repeat protein kinase family protein |
-0.217 |
-1.565 |
| 61 |
256671_at |
AT3G52290
|
IQD3, IQ-domain 3 |
-0.212 |
-1.039 |
| 62 |
256138_at |
AT1G48670
|
[auxin-responsive GH3 family protein] |
-0.212 |
-0.306 |
| 63 |
258988_at |
AT3G08890
|
unknown |
-0.207 |
1.217 |
| 64 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.205 |
-1.989 |
| 65 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
-0.203 |
-0.966 |
| 66 |
261175_at |
AT1G04800
|
[glycine-rich protein] |
-0.202 |
-0.802 |
| 67 |
248073_at |
AT5G55720
|
[Pectin lyase-like superfamily protein] |
-0.199 |
0.218 |
| 68 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.199 |
-1.752 |
| 69 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.192 |
-1.015 |