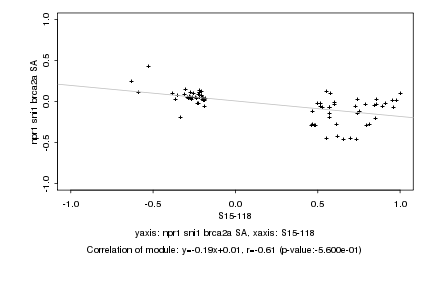
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
S15-118 |
Log2 signal ratio
npr1 sni1 brca2a SA |
| 1 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
1.000 |
0.107 |
| 2 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.976 |
0.019 |
| 3 |
257473_at |
AT1G33840
|
unknown |
0.957 |
-0.065 |
| 4 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.952 |
0.021 |
| 5 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.911 |
-0.015 |
| 6 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.893 |
-0.054 |
| 7 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.854 |
-0.030 |
| 8 |
263851_at |
AT2G04460
|
unknown |
0.851 |
0.025 |
| 9 |
258203_at |
AT3G13950
|
unknown |
0.850 |
-0.201 |
| 10 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.839 |
-0.047 |
| 11 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.810 |
-0.273 |
| 12 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.795 |
-0.282 |
| 13 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.787 |
-0.032 |
| 14 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.751 |
-0.112 |
| 15 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.741 |
0.029 |
| 16 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.738 |
-0.136 |
| 17 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.731 |
-0.457 |
| 18 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.726 |
-0.056 |
| 19 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.695 |
-0.439 |
| 20 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
0.654 |
-0.459 |
| 21 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.619 |
-0.422 |
| 22 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.609 |
-0.275 |
| 23 |
264635_at |
AT1G65500
|
unknown |
0.601 |
-0.004 |
| 24 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.596 |
-0.034 |
| 25 |
263515_at |
AT2G21640
|
unknown |
0.573 |
0.102 |
| 26 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.570 |
-0.145 |
| 27 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.570 |
-0.184 |
| 28 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.568 |
-0.062 |
| 29 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.566 |
-0.061 |
| 30 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.550 |
-0.447 |
| 31 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.547 |
0.132 |
| 32 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.524 |
-0.071 |
| 33 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.514 |
-0.053 |
| 34 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.511 |
-0.016 |
| 35 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.494 |
-0.017 |
| 36 |
266017_at |
AT2G18690
|
unknown |
0.484 |
-0.290 |
| 37 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.477 |
-0.286 |
| 38 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.467 |
-0.121 |
| 39 |
262085_at |
AT1G56060
|
unknown |
0.462 |
-0.278 |
| 40 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
0.460 |
-0.287 |
| 41 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.634 |
0.250 |
| 42 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
-0.594 |
0.122 |
| 43 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.528 |
0.432 |
| 44 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.388 |
0.104 |
| 45 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.367 |
0.028 |
| 46 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.352 |
0.081 |
| 47 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.337 |
-0.186 |
| 48 |
250753_at |
AT5G05860
|
UGT76C2, UDP-glucosyl transferase 76C2 |
-0.312 |
0.095 |
| 49 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.306 |
0.156 |
| 50 |
258392_at |
AT3G15400
|
ATA20, anther 20 |
-0.295 |
0.057 |
| 51 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.286 |
0.038 |
| 52 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
-0.279 |
0.071 |
| 53 |
258888_at |
AT3G05620
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.277 |
0.054 |
| 54 |
263002_at |
AT1G54200
|
unknown |
-0.273 |
0.114 |
| 55 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.270 |
0.036 |
| 56 |
250281_at |
AT5G13240
|
[transcription regulators] |
-0.262 |
0.055 |
| 57 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.259 |
0.105 |
| 58 |
255637_at |
AT4G00750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.245 |
0.055 |
| 59 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.240 |
0.039 |
| 60 |
258990_at |
AT3G08840
|
[D-alanine--D-alanine ligase family] |
-0.236 |
-0.023 |
| 61 |
261097_at |
AT1G62950
|
[leucine-rich repeat transmembrane protein kinase family protein] |
-0.227 |
0.093 |
| 62 |
256138_at |
AT1G48670
|
[auxin-responsive GH3 family protein] |
-0.227 |
-0.016 |
| 63 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.224 |
0.111 |
| 64 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.222 |
0.051 |
| 65 |
251219_at |
AT3G62390
|
TBL6, TRICHOME BIREFRINGENCE-LIKE 6 |
-0.220 |
0.114 |
| 66 |
265118_at |
AT1G62660
|
[Glycosyl hydrolases family 32 protein] |
-0.220 |
0.135 |
| 67 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.212 |
0.074 |
| 68 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.209 |
0.129 |
| 69 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
-0.207 |
0.076 |
| 70 |
262090_at |
AT1G55970
|
HAC04, HAC4, histone acetyltransferase of the CBP family 4, HAC6, HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY 6, HAG04, HISTONE ACETYLTRANSFERASE OF THE GNAT/MYST SUPERFAMILY 04, HAG4, HISTONE ACETYLTRANSFERASE OF THE GNAT/MYST SUPERFAMILY 4 |
-0.201 |
0.036 |
| 71 |
259843_at |
AT1G73570
|
[HCP-like superfamily protein] |
-0.201 |
0.059 |
| 72 |
245834_at |
AT1G42200
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.3e-27 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.200 |
0.041 |
| 73 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
-0.198 |
0.021 |
| 74 |
254791_at |
AT4G12910
|
scpl20, serine carboxypeptidase-like 20 |
-0.197 |
0.036 |
| 75 |
263748_at |
AT2G21480
|
[Malectin/receptor-like protein kinase family protein] |
-0.195 |
0.013 |
| 76 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
-0.192 |
0.027 |
| 77 |
265949_at |
AT2G18540
|
[RmlC-like cupins superfamily protein] |
-0.192 |
-0.052 |
| 78 |
264962_at |
AT1G77110
|
PIN6, PIN-FORMED 6 |
-0.190 |
0.013 |
| 79 |
252747_at |
AT3G43320
|
unknown |
-0.183 |
0.044 |