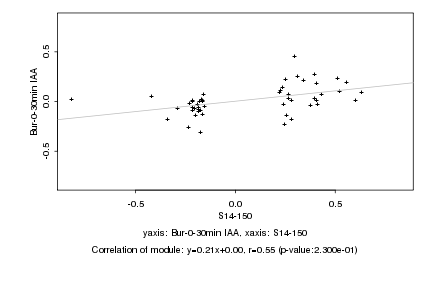
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
S14-150 |
Log2 signal ratio
Bur-0-30min IAA |
| 1 |
263515_at |
AT2G21640
|
unknown |
0.630 |
0.098 |
| 2 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.598 |
0.017 |
| 3 |
260522_x_at |
AT2G41730
|
unknown |
0.554 |
0.195 |
| 4 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.519 |
0.105 |
| 5 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.511 |
0.233 |
| 6 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.428 |
0.076 |
| 7 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.410 |
-0.021 |
| 8 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.402 |
0.186 |
| 9 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
0.402 |
0.020 |
| 10 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.393 |
0.037 |
| 11 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
0.393 |
0.280 |
| 12 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.372 |
-0.040 |
| 13 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.337 |
0.219 |
| 14 |
266017_at |
AT2G18690
|
unknown |
0.309 |
0.256 |
| 15 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.293 |
0.460 |
| 16 |
245401_at |
AT4G17670
|
unknown |
0.280 |
0.017 |
| 17 |
262388_at |
AT1G49320
|
unknown |
0.277 |
-0.171 |
| 18 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.265 |
0.034 |
| 19 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.262 |
0.075 |
| 20 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.254 |
-0.133 |
| 21 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.247 |
0.223 |
| 22 |
252536_at |
AT3G45700
|
[Major facilitator superfamily protein] |
0.241 |
-0.229 |
| 23 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.239 |
-0.029 |
| 24 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.235 |
0.147 |
| 25 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.223 |
0.119 |
| 26 |
261193_at |
AT1G32920
|
unknown |
0.216 |
0.092 |
| 27 |
247564_at |
AT5G61140
|
[U5 small nuclear ribonucleoprotein helicase] |
-0.825 |
0.025 |
| 28 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.424 |
0.059 |
| 29 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
-0.342 |
-0.179 |
| 30 |
256367_at |
AT1G66810
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
-0.291 |
-0.061 |
| 31 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.239 |
-0.257 |
| 32 |
265103_at |
AT1G31070
|
GlcNAc1pUT1, N-acetylglucosamine-1-phosphate uridylyltransferase 1 |
-0.233 |
-0.012 |
| 33 |
252836_at |
AT3G42070
|
unknown |
-0.220 |
0.009 |
| 34 |
259530_at |
AT1G12450
|
[SNARE associated Golgi protein family] |
-0.218 |
-0.055 |
| 35 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.216 |
0.020 |
| 36 |
250281_at |
AT5G13240
|
[transcription regulators] |
-0.216 |
-0.085 |
| 37 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
-0.210 |
-0.066 |
| 38 |
259203_at |
AT3G09130
|
unknown |
-0.201 |
-0.131 |
| 39 |
250375_at |
AT5G11540
|
AtGulLO3, GulLO3, L -gulono-1,4-lactone ( L -GulL) oxidase 3 |
-0.191 |
-0.021 |
| 40 |
248073_at |
AT5G55720
|
[Pectin lyase-like superfamily protein] |
-0.188 |
-0.093 |
| 41 |
253320_at |
AT4G34020
|
AtDJ1C, DJ-1 homolog C, DJ-1c, DJ1C, DJ-1 homolog C |
-0.186 |
-0.055 |
| 42 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
-0.186 |
-0.073 |
| 43 |
254322_at |
AT4G22600
|
INP1, INAPERTURATE POLLEN1 |
-0.185 |
0.007 |
| 44 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
-0.178 |
-0.082 |
| 45 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
-0.176 |
-0.302 |
| 46 |
247773_at |
AT5G58630
|
TRM31, TON1 Recruiting Motif 31 |
-0.171 |
0.024 |
| 47 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.169 |
0.020 |
| 48 |
248468_at |
AT5G50750
|
RGP4, reversibly glycosylated polypeptide 4 |
-0.167 |
0.007 |
| 49 |
260573_at |
AT2G47280
|
[Pectin lyase-like superfamily protein] |
-0.166 |
-0.130 |
| 50 |
248488_at |
AT5G51080
|
[RNase H family protein] |
-0.164 |
0.074 |
| 51 |
250766_at |
AT5G05550
|
[sequence-specific DNA binding transcription factors] |
-0.160 |
-0.049 |