|
probeID |
AGICode |
Annotation |
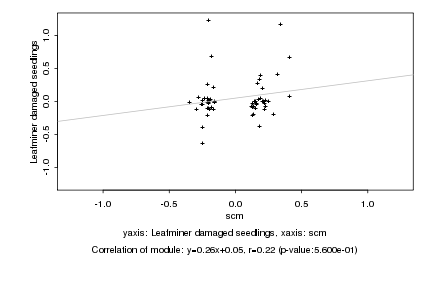
Log2 signal ratio
scm |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
264892_at |
AT1G23160
|
[Auxin-responsive GH3 family protein] |
0.404 |
0.078 |
| 2 |
257466_at |
AT1G62840
|
unknown |
0.401 |
0.680 |
| 3 |
245209_at |
AT5G12340
|
unknown |
0.339 |
1.171 |
| 4 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.312 |
0.416 |
| 5 |
267367_at |
AT2G44210
|
unknown |
0.284 |
-0.195 |
| 6 |
255022_at |
AT4G10350
|
ANAC070, NAC domain containing protein 70, BRN2, BEARSKIN 2, NAC070, NAC domain containing protein 70 |
0.249 |
0.011 |
| 7 |
267420_at |
AT2G35030
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.227 |
0.021 |
| 8 |
266539_at |
AT2G35160
|
SGD9, SET DOMAIN-CONTAINING PROTEIN 9, SUVH5, SU(VAR)3-9 homolog 5 |
0.220 |
-0.067 |
| 9 |
248425_at |
AT5G51690
|
ACS12, 1-amino-cyclopropane-1-carboxylate synthase 12 |
0.219 |
-0.112 |
| 10 |
254802_at |
AT4G13090
|
XTH2, xyloglucan endotransglucosylase/hydrolase 2 |
0.217 |
-0.023 |
| 11 |
257358_at |
AT2G40990
|
[DHHC-type zinc finger family protein] |
0.206 |
0.014 |
| 12 |
255859_at |
AT5G34930
|
[arogenate dehydrogenase] |
0.203 |
0.207 |
| 13 |
248394_at |
AT5G52070
|
[Agenet domain-containing protein] |
0.203 |
0.012 |
| 14 |
254917_at |
AT4G11350
|
unknown |
0.186 |
0.406 |
| 15 |
256063_at |
AT1G07130
|
ATSTN1, STN1 |
0.184 |
0.056 |
| 16 |
254924_at |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
0.181 |
0.337 |
| 17 |
260363_at |
AT1G70550
|
unknown |
0.179 |
-0.371 |
| 18 |
266976_at |
AT2G39410
|
[alpha/beta-Hydrolases superfamily protein] |
0.171 |
0.039 |
| 19 |
252484_at |
AT3G46690
|
[UDP-Glycosyltransferase superfamily protein] |
0.160 |
0.285 |
| 20 |
249470_at |
AT5G39350
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.159 |
-0.044 |
| 21 |
246869_at |
AT5G26020
|
[similar to Glutamic acid-rich protein precursor (GB:P13816)] |
0.155 |
-0.020 |
| 22 |
258627_at |
AT3G04460
|
APM4, ABERRANT PEROXISOME MORPHOLOGY 4, ATPEX12, PEROXIN-12, PEX12, peroxin-12 |
0.147 |
-0.091 |
| 23 |
245157_at |
AT2G33160
|
NMA, NIMNA (Sanskrit for 'sunken or low) |
0.145 |
0.015 |
| 24 |
253020_at |
AT4G38020
|
[tRNA/rRNA methyltransferase (SpoU) family protein] |
0.137 |
0.001 |
| 25 |
265688_at |
AT2G24300
|
[Calmodulin-binding protein] |
0.132 |
-0.066 |
| 26 |
265986_at |
AT2G24230
|
[Leucine-rich repeat protein kinase family protein] |
0.131 |
-0.183 |
| 27 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
0.126 |
-0.207 |
| 28 |
251693_at |
AT3G56550
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.123 |
-0.026 |
| 29 |
245509_at |
AT4G15730
|
[CW-type Zinc Finger] |
0.122 |
-0.089 |
| 30 |
245926_at |
AT5G24740
|
SHBY, SHRUBBY |
0.121 |
-0.065 |
| 31 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
-0.354 |
-0.013 |
| 32 |
250812_at |
AT5G04900
|
NOL, NYC1-like |
-0.299 |
-0.117 |
| 33 |
262578_at |
AT1G15300
|
CYTOSLEEPER, CYTOSLEEPER |
-0.284 |
0.072 |
| 34 |
245620_at |
AT4G14050
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.257 |
-0.040 |
| 35 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.255 |
-0.379 |
| 36 |
266387_at |
AT2G32360
|
[Ubiquitin-like superfamily protein] |
-0.255 |
-0.031 |
| 37 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.254 |
0.030 |
| 38 |
263674_at |
AT2G04790
|
unknown |
-0.253 |
-0.631 |
| 39 |
266492_at |
AT2G07020
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.236 |
0.052 |
| 40 |
245933_at |
AT5G09320
|
VPS9B |
-0.216 |
-0.097 |
| 41 |
254948_at |
AT4G11000
|
[Ankyrin repeat family protein] |
-0.215 |
0.263 |
| 42 |
262907_at |
AT1G59720
|
CRR28, CHLORORESPIRATORY REDUCTION28 |
-0.214 |
-0.207 |
| 43 |
254406_at |
AT4G21360
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
-0.213 |
0.047 |
| 44 |
262190_at |
AT1G78030
|
unknown |
-0.212 |
-0.002 |
| 45 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.211 |
-0.093 |
| 46 |
255173_at |
AT4G08020
|
[CACTA-like transposase family (En/Spm), has a 2.5e-93 P-value blast match to GB:BAA20532 ORF of transposon Tdc1 (CACTA-element) (Daucus carota)] |
-0.210 |
-0.027 |
| 47 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.208 |
1.244 |
| 48 |
265086_at |
AT1G03770
|
ATRING1B, ARABIDOPSIS THALIANA RING 1B, RING1B, RING 1B |
-0.208 |
0.030 |
| 49 |
256083_at |
AT1G20735
|
[F-box and associated interaction domains-containing protein] |
-0.205 |
-0.010 |
| 50 |
247714_at |
AT5G59340
|
WOX2, WUSCHEL related homeobox 2 |
-0.201 |
-0.116 |
| 51 |
248969_at |
AT5G45310
|
unknown |
-0.196 |
0.028 |
| 52 |
251540_at |
AT3G58740
|
CSY1, citrate synthase 1 |
-0.192 |
0.035 |
| 53 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.184 |
0.698 |
| 54 |
252809_at |
AT3G42430
|
unknown |
-0.183 |
-0.077 |
| 55 |
261445_at |
AT1G28380
|
NSL1, necrotic spotted lesions 1 |
-0.172 |
0.224 |
| 56 |
249386_at |
AT5G40060
|
[Disease resistance protein (NBS-LRR class) family] |
-0.167 |
-0.106 |
| 57 |
263354_at |
AT2G22060
|
unknown |
-0.163 |
-0.012 |
| 58 |
255352_at |
AT4G03860
|
[transposon protein -related, similar to athila retroelement] |
-0.161 |
-0.006 |
| 59 |
255574_at |
AT4G01420
|
CBL5, calcineurin B-like protein 5 |
-0.160 |
0.007 |