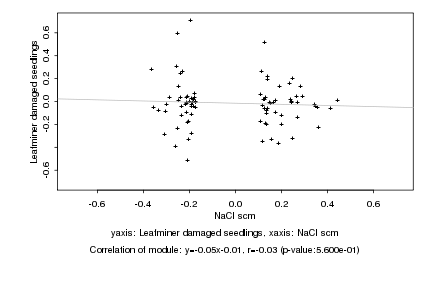
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
NaCl scm |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
257143_at |
AT3G20110
|
CYP705A20, cytochrome P450, family 705, subfamily A, polypeptide 20 |
0.441 |
0.014 |
| 2 |
262305_at |
AT1G70950
|
[TPX2 (targeting protein for Xklp2) protein family] |
0.413 |
-0.053 |
| 3 |
265463_at |
AT2G37090
|
IRX9, IRREGULAR XYLEM 9 |
0.361 |
-0.225 |
| 4 |
267511_at |
AT2G45670
|
[calcineurin B subunit-related] |
0.355 |
-0.047 |
| 5 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
0.346 |
-0.035 |
| 6 |
253211_at |
AT4G34880
|
[Amidase family protein] |
0.340 |
-0.026 |
| 7 |
251875_at |
AT3G54270
|
[sucrose-6F-phosphate phosphohydrolase family protein] |
0.291 |
0.052 |
| 8 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
0.281 |
0.136 |
| 9 |
253098_at |
AT4G37340
|
CYP81D3, cytochrome P450, family 81, subfamily D, polypeptide 3 |
0.266 |
-0.002 |
| 10 |
260113_at |
AT1G63300
|
[Myosin heavy chain-related protein] |
0.266 |
-0.131 |
| 11 |
253122_at |
AT4G35987
|
CaM KMT, calmodulin N-methyltransferase |
0.262 |
0.048 |
| 12 |
257593_at |
AT3G24840
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.245 |
0.203 |
| 13 |
266715_at |
AT2G46780
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.245 |
-0.316 |
| 14 |
260987_at |
AT1G53590
|
NTMC2T6.1, NTMC2TYPE6.1 |
0.241 |
0.005 |
| 15 |
262220_at |
AT1G74740
|
ATCPK30, CDPK1A, CALCIUM-DEPENDENT PROTEIN KINASE 1A, CPK30, calcium-dependent protein kinase 30 |
0.240 |
-0.005 |
| 16 |
253782_at |
AT4G28590
|
AtECB1, ECB1, early chloroplast biogenesis 1, MRL7, Mesophyll-cell RNAi Library line 7, SVR4, SUPPRESSOR OF VARIEGATION 4 |
0.238 |
0.018 |
| 17 |
258046_at |
AT3G21220
|
ATMAP2K_ALPHA, ATMEK5, ARABIDOPSIS THALIANA MITOGEN-ACTIVATED PROTEIN KINASE KINASE 5, ATMKK5, ARABIDOPSIS THALIANA MITOGEN-ACTIVATED PROTEIN KINASE KINASE 5, MAP2K_A, MEK5, MAP KINASE KINASE 5, MKK5, MAP kinase kinase 5 |
0.234 |
0.161 |
| 18 |
253034_at |
AT4G38230
|
ATCPK26, ARABIDOPSIS THALIANA CALCIUM-DEPENDENT PROTEIN KINASE 26, CPK26, calcium-dependent protein kinase 26 |
0.200 |
-0.119 |
| 19 |
256950_at |
AT3G19080
|
[SWIB complex BAF60b domain-containing protein] |
0.198 |
-0.199 |
| 20 |
246614_at |
AT5G35410
|
ATSOS2, SALT OVERLY SENSITIVE 2, CIPK24, CBL-INTERACTING PROTEIN KINASE 24, SNRK3.11, SNF1-RELATED PROTEIN KINASE 3.11, SOS2, SALT OVERLY SENSITIVE 2 |
0.191 |
0.137 |
| 21 |
264700_at |
AT1G70100
|
unknown |
0.186 |
-0.358 |
| 22 |
246277_at |
AT4G36460
|
unknown |
0.173 |
0.016 |
| 23 |
245055_at |
AT2G26470
|
unknown |
0.171 |
-0.089 |
| 24 |
250913_at |
AT5G03770
|
AtKdtA, KDTA, KDO transferase A |
0.163 |
0.000 |
| 25 |
261363_at |
AT1G41830
|
SKS6, SKU5 SIMILAR 6, SKS6, SKU5-similar 6 |
0.156 |
-0.324 |
| 26 |
251887_at |
AT3G54170
|
ATFIP37, FKBP12 interacting protein 37, FIP37, FKBP12 interacting protein 37 |
0.152 |
-0.014 |
| 27 |
260021_at |
AT1G30010
|
nMAT1, nuclear maturase 1 |
0.145 |
-0.004 |
| 28 |
246529_at |
AT5G15730
|
AtCRLK2, CRLK2, calcium/calmodulin-regulated receptor-like kinase 2 |
0.139 |
0.195 |
| 29 |
262410_at |
AT1G34770
|
unknown |
0.137 |
-0.054 |
| 30 |
261125_at |
AT1G04990
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.137 |
0.226 |
| 31 |
266713_at |
AT2G46760
|
AtGulLO6, GulLO6, L -gulono-1,4-lactone ( L -GulL) oxidase 6 |
0.133 |
-0.099 |
| 32 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
0.133 |
-0.200 |
| 33 |
254860_at |
AT4G12110
|
ATSMO1-1, SMO1-1, sterol-4alpha-methyl oxidase 1-1 |
0.132 |
-0.071 |
| 34 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
0.128 |
-0.184 |
| 35 |
245280_at |
AT4G16845
|
VRN2, REDUCED VERNALIZATION RESPONSE 2 |
0.127 |
0.042 |
| 36 |
256137_at |
AT1G48690
|
[Auxin-responsive GH3 family protein] |
0.126 |
-0.056 |
| 37 |
244931_at |
ATMG00630
|
ORF110B |
0.125 |
0.020 |
| 38 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.123 |
0.518 |
| 39 |
249603_at |
AT5G37210
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.122 |
0.025 |
| 40 |
249189_at |
AT5G42780
|
AtHB27, homeobox protein 27, HB27, homeobox protein 27, ZHD13, ZINC FINGER HOMEODOMAIN 13 |
0.116 |
-0.347 |
| 41 |
256132_at |
AT1G13610
|
[alpha/beta-Hydrolases superfamily protein] |
0.114 |
-0.034 |
| 42 |
252822_at |
AT4G39955
|
[alpha/beta-Hydrolases superfamily protein] |
0.111 |
0.268 |
| 43 |
265585_at |
AT2G20000
|
CDC27b, HBT, HOBBIT |
0.107 |
-0.170 |
| 44 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
0.106 |
0.067 |
| 45 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
-0.367 |
0.287 |
| 46 |
256584_at |
AT3G28750
|
unknown |
-0.360 |
-0.052 |
| 47 |
257028_at |
AT3G19230
|
[Leucine-rich repeat (LRR) family protein] |
-0.335 |
-0.073 |
| 48 |
255277_at |
AT4G04890
|
PDF2, protodermal factor 2 |
-0.309 |
-0.285 |
| 49 |
246664_at |
AT5G34800
|
[pseudogene] |
-0.305 |
-0.086 |
| 50 |
263052_at |
AT2G13430
|
unknown |
-0.304 |
-0.024 |
| 51 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
-0.291 |
0.043 |
| 52 |
260769_at |
AT1G49010
|
[Duplicated homeodomain-like superfamily protein] |
-0.264 |
-0.385 |
| 53 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
-0.258 |
0.306 |
| 54 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.256 |
0.596 |
| 55 |
256860_at |
AT3G23840
|
CER26-LIKE, CER26-LIKE |
-0.254 |
-0.233 |
| 56 |
249586_at |
AT5G37840
|
unknown |
-0.249 |
0.133 |
| 57 |
256924_at |
AT3G29590
|
AT5MAT |
-0.248 |
0.013 |
| 58 |
263197_at |
AT1G53930
|
[Ubiquitin-like superfamily protein] |
-0.243 |
0.040 |
| 59 |
251916_at |
AT3G53960
|
[Major facilitator superfamily protein] |
-0.241 |
0.246 |
| 60 |
264480_at |
AT1G77300
|
ASHH2, ASH1 HOMOLOG 2, CCR1, CAROTENOID CHLOROPLAST REGULATORY1, EFS, EARLY FLOWERING IN SHORT DAYS, LAZ2, LAZARUS 2, SDG8, SET DOMAIN GROUP 8 |
-0.237 |
-0.039 |
| 61 |
265998_at |
AT2G24270
|
ALDH11A3, aldehyde dehydrogenase 11A3 |
-0.236 |
-0.121 |
| 62 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.231 |
0.266 |
| 63 |
245992_at |
AT5G20690
|
PRK6, pollen receptor like kinase 6 |
-0.219 |
-0.022 |
| 64 |
247226_at |
AT5G65100
|
[Ethylene insensitive 3 family protein] |
-0.216 |
-0.001 |
| 65 |
252835_at |
AT3G42060
|
[myosin heavy chain-related] |
-0.215 |
-0.016 |
| 66 |
257196_at |
AT3G23790
|
AAE16, acyl activating enzyme 16 |
-0.215 |
0.039 |
| 67 |
265403_at |
AT2G16690
|
unknown |
-0.214 |
-0.090 |
| 68 |
254212_at |
AT4G23580
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.210 |
0.044 |
| 69 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.209 |
-0.179 |
| 70 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.209 |
-0.513 |
| 71 |
254319_at |
AT4G22560
|
unknown |
-0.208 |
-0.324 |
| 72 |
256942_at |
AT3G23290
|
LSH4, LIGHT SENSITIVE HYPOCOTYLS 4 |
-0.207 |
-0.167 |
| 73 |
260019_at |
AT1G41825
|
[gypsy-like retrotransposon family (Athila), has a 1.9e-98 P-value blast match to gb|AAL06422.1|AF378081_1 reverse transcriptase (Athila4) (Arabidopsis thaliana) (Gypsy_Ty3-family)] |
-0.203 |
0.007 |
| 74 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-0.197 |
0.716 |
| 75 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.195 |
-0.112 |
| 76 |
252263_at |
AT3G49520
|
[F-box and associated interaction domains-containing protein] |
-0.194 |
-0.019 |
| 77 |
264684_at |
AT1G65590
|
ATHEX1, HEXO3, beta-hexosaminidase 3 |
-0.192 |
-0.272 |
| 78 |
258590_at |
AT3G04280
|
ARR22, response regulator 22, RR22, response regulator 22 |
-0.192 |
-0.040 |
| 79 |
265261_at |
AT2G42990
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.192 |
0.035 |
| 80 |
251469_at |
AT3G59530
|
LAP3, LESS ADHERENT POLLEN 3 |
-0.187 |
-0.002 |
| 81 |
257698_at |
AT3G12730
|
[Homeodomain-like superfamily protein] |
-0.185 |
0.018 |
| 82 |
261441_at |
AT1G28470
|
ANAC010, NAC domain containing protein 10, NAC010, NAC domain containing protein 10, SND3, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 3 |
-0.184 |
-0.036 |
| 83 |
253902_at |
AT4G27170
|
AT2S4, SESA4, seed storage albumin 4 |
-0.182 |
0.040 |
| 84 |
250464_at |
AT5G10040
|
unknown |
-0.181 |
0.076 |
| 85 |
265997_at |
AT2G24250
|
unknown |
-0.178 |
-0.049 |
| 86 |
254294_at |
AT4G23070
|
ATRBL7, RHOMBOID-like protein 7, RBL7, RHOMBOID-like protein 7 |
-0.178 |
0.000 |
| 87 |
255741_at |
AT1G25410
|
ATIPT6, ARABIDOPSIS THALIANA ISOPENTENYLTRANSFERASE 6, IPT6, isopentenyltransferase 6 |
-0.174 |
0.003 |