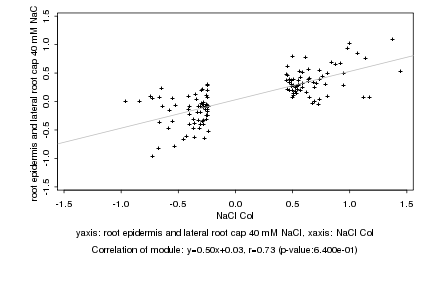
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
NaCl Col |
Log2 signal ratio
root epidermis and lateral root cap 40 mM NaCl |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.441 |
0.543 |
| 2 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
1.371 |
1.100 |
| 3 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
1.172 |
0.076 |
| 4 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.132 |
0.757 |
| 5 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
1.114 |
0.088 |
| 6 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.065 |
0.847 |
| 7 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.993 |
1.032 |
| 8 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.980 |
0.936 |
| 9 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.939 |
0.499 |
| 10 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.939 |
0.297 |
| 11 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.915 |
0.671 |
| 12 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.873 |
0.665 |
| 13 |
265024_at |
AT1G24600
|
unknown |
0.832 |
0.699 |
| 14 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
0.801 |
0.096 |
| 15 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.797 |
0.501 |
| 16 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.780 |
0.313 |
| 17 |
263881_at |
AT2G21820
|
unknown |
0.760 |
0.448 |
| 18 |
248209_at |
AT5G53990
|
[UDP-Glycosyltransferase superfamily protein] |
0.733 |
0.042 |
| 19 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
0.731 |
0.548 |
| 20 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.727 |
0.400 |
| 21 |
265983_at |
AT2G18550
|
ATHB21, homeobox protein 21, HB-2, homeobox-2, HB21, homeobox protein 21 |
0.726 |
-0.036 |
| 22 |
266274_at |
AT2G29380
|
HAI3, highly ABA-induced PP2C gene 3 |
0.701 |
0.327 |
| 23 |
250158_at |
AT5G15190
|
unknown |
0.684 |
0.252 |
| 24 |
252073_at |
AT3G51750
|
unknown |
0.684 |
0.008 |
| 25 |
249771_at |
AT5G24080
|
[Protein kinase superfamily protein] |
0.677 |
0.343 |
| 26 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
0.667 |
-0.028 |
| 27 |
246582_at |
AT1G31750
|
[proline-rich family protein] |
0.645 |
0.406 |
| 28 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.642 |
0.085 |
| 29 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.639 |
0.567 |
| 30 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.632 |
0.366 |
| 31 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.631 |
0.400 |
| 32 |
253936_at |
AT4G26880
|
[Stigma-specific Stig1 family protein] |
0.620 |
0.175 |
| 33 |
251644_at |
AT3G57540
|
[Remorin family protein] |
0.609 |
0.777 |
| 34 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.583 |
0.250 |
| 35 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.582 |
0.325 |
| 36 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
0.581 |
0.518 |
| 37 |
251132_at |
AT5G01200
|
[Duplicated homeodomain-like superfamily protein] |
0.569 |
0.199 |
| 38 |
262061_at |
AT1G80110
|
ATPP2-B11, phloem protein 2-B11, PP2-B11, phloem protein 2-B11 |
0.566 |
0.439 |
| 39 |
267058_at |
AT2G32510
|
MAPKKK17, mitogen-activated protein kinase kinase kinase 17 |
0.556 |
0.534 |
| 40 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
0.550 |
0.316 |
| 41 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.546 |
0.379 |
| 42 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.544 |
0.287 |
| 43 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.537 |
0.216 |
| 44 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.534 |
0.266 |
| 45 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.531 |
0.155 |
| 46 |
248764_at |
AT5G47640
|
NF-YB2, nuclear factor Y, subunit B2 |
0.524 |
0.247 |
| 47 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
0.520 |
0.185 |
| 48 |
250858_at |
AT5G04760
|
[Duplicated homeodomain-like superfamily protein] |
0.508 |
0.282 |
| 49 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.506 |
0.111 |
| 50 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.506 |
0.398 |
| 51 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.502 |
0.158 |
| 52 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.494 |
0.076 |
| 53 |
260243_at |
AT1G63720
|
unknown |
0.492 |
0.194 |
| 54 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.491 |
0.799 |
| 55 |
265204_at |
AT2G36650
|
unknown |
0.486 |
0.272 |
| 56 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
0.486 |
0.304 |
| 57 |
245724_at |
AT1G73390
|
[Endosomal targeting BRO1-like domain-containing protein] |
0.484 |
0.377 |
| 58 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.472 |
0.394 |
| 59 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
0.471 |
0.344 |
| 60 |
247120_at |
AT5G65990
|
[Transmembrane amino acid transporter family protein] |
0.468 |
0.211 |
| 61 |
262496_at |
AT1G21790
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
0.465 |
0.340 |
| 62 |
256114_at |
AT1G16850
|
unknown |
0.454 |
0.459 |
| 63 |
258878_at |
AT3G03170
|
unknown |
0.449 |
0.218 |
| 64 |
262363_at |
AT1G72850
|
[Disease resistance protein (TIR-NBS class)] |
0.448 |
0.619 |
| 65 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.444 |
0.377 |
| 66 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.443 |
0.475 |
| 67 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.963 |
0.007 |
| 68 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
-0.845 |
0.008 |
| 69 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.746 |
0.093 |
| 70 |
251368_at |
AT3G61380
|
TRM14, TON1 Recruiting Motif 14 |
-0.732 |
0.069 |
| 71 |
252511_at |
AT3G46280
|
[protein kinase-related] |
-0.731 |
-0.966 |
| 72 |
255608_at |
AT4G01140
|
unknown |
-0.680 |
-0.823 |
| 73 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.673 |
0.074 |
| 74 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.671 |
-0.362 |
| 75 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
-0.653 |
0.233 |
| 76 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.640 |
-0.070 |
| 77 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.593 |
-0.470 |
| 78 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.580 |
-0.155 |
| 79 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.551 |
-0.346 |
| 80 |
264947_at |
AT1G77020
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.551 |
0.060 |
| 81 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.542 |
-0.785 |
| 82 |
248443_at |
AT5G51310
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.529 |
-0.067 |
| 83 |
267240_at |
AT2G02680
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.458 |
-0.664 |
| 84 |
246997_at |
AT5G67390
|
unknown |
-0.437 |
-0.604 |
| 85 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.413 |
0.090 |
| 86 |
251688_at |
AT3G56480
|
[myosin heavy chain-related] |
-0.413 |
-0.138 |
| 87 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.410 |
-0.085 |
| 88 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.406 |
-0.220 |
| 89 |
252582_at |
AT3G45530
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.405 |
-0.395 |
| 90 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.369 |
-0.466 |
| 91 |
254487_at |
AT4G20780
|
CML42, calmodulin like 42 |
-0.368 |
-0.300 |
| 92 |
245574_at |
AT4G14750
|
IQD19, IQ-domain 19 |
-0.362 |
-0.624 |
| 93 |
259664_at |
AT1G55330
|
AGP21, arabinogalactan protein 21, ATAGP21, ARABINOGALACTAN PROTEIN 21 |
-0.362 |
-0.384 |
| 94 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
-0.350 |
0.128 |
| 95 |
249435_at |
AT5G39970
|
[catalytics] |
-0.348 |
0.037 |
| 96 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
-0.338 |
-0.178 |
| 97 |
262793_at |
AT1G13110
|
CYP71B7, cytochrome P450, family 71 subfamily B, polypeptide 7 |
-0.330 |
-0.324 |
| 98 |
262772_at |
AT1G13210
|
ACA.l, autoinhibited Ca2+/ATPase II |
-0.330 |
-0.075 |
| 99 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.316 |
-0.465 |
| 100 |
259888_at |
AT1G76350
|
NLP5, NIN-like protein 5 |
-0.314 |
-0.184 |
| 101 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
-0.313 |
-0.078 |
| 102 |
252084_at |
AT3G51970
|
ASAT1, acyl-CoA sterol acyl transferase 1, ATASAT1, ATSAT1, ARABIDOPSIS THALIANA STEROL O-ACYLTRANSFERASE 1 |
-0.310 |
-0.393 |
| 103 |
246898_at |
AT5G25580
|
unknown |
-0.305 |
-0.026 |
| 104 |
254344_at |
AT4G22110
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.302 |
0.194 |
| 105 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
-0.295 |
0.226 |
| 106 |
251857_at |
AT3G54770
|
ARP1, ABA-regulated RNA-binding protein 1 |
-0.292 |
-0.336 |
| 107 |
249166_at |
AT5G42840
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.290 |
-0.041 |
| 108 |
246797_at |
AT5G26790
|
unknown |
-0.285 |
-0.009 |
| 109 |
264481_at |
AT1G77200
|
[Integrase-type DNA-binding superfamily protein] |
-0.284 |
-0.329 |
| 110 |
250695_at |
AT5G06740
|
LecRK-S.5, L-type lectin receptor kinase S.5 |
-0.282 |
-0.044 |
| 111 |
251944_at |
AT3G53510
|
ABCG20, ATP-binding cassette G20 |
-0.282 |
-0.104 |
| 112 |
249768_at |
AT5G24100
|
[Leucine-rich repeat protein kinase family protein] |
-0.280 |
-0.390 |
| 113 |
254193_at |
AT4G23870
|
unknown |
-0.276 |
-0.636 |
| 114 |
265335_at |
AT2G18245
|
[alpha/beta-Hydrolases superfamily protein] |
-0.270 |
-0.070 |
| 115 |
261772_at |
AT1G76240
|
unknown |
-0.269 |
-0.104 |
| 116 |
260432_at |
AT1G68150
|
ATWRKY9, WRKY9, WRKY DNA-binding protein 9 |
-0.267 |
-0.126 |
| 117 |
250823_at |
AT5G05180
|
unknown |
-0.262 |
-0.048 |
| 118 |
247483_at |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.256 |
0.112 |
| 119 |
251048_at |
AT5G02410
|
ALG10, homolog of yeast ALG10 |
-0.255 |
-0.314 |
| 120 |
259919_at |
AT1G72560
|
PSD, PAUSED |
-0.255 |
-0.243 |
| 121 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
-0.253 |
-0.245 |
| 122 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
-0.252 |
0.315 |
| 123 |
254060_at |
AT4G25350
|
SHB1, SHORT HYPOCOTYL UNDER BLUE1 |
-0.248 |
0.087 |
| 124 |
246315_at |
AT3G56870
|
unknown |
-0.247 |
-0.167 |
| 125 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
-0.247 |
0.296 |
| 126 |
262170_at |
AT1G74940
|
unknown |
-0.247 |
-0.123 |
| 127 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
-0.246 |
0.204 |
| 128 |
256754_at |
AT3G25690
|
AtCHUP1, Arabidopsis thaliana CHLOROPLAST UNUSUAL POSITIONING 1, CHUP1, CHLOROPLAST UNUSUAL POSITIONING 1 |
-0.245 |
-0.055 |
| 129 |
264770_at |
AT1G23030
|
[ARM repeat superfamily protein] |
-0.245 |
-0.236 |
| 130 |
264835_at |
AT1G03550
|
AtSCAMP4, SCAMP4, Secretory carrier membrane protein 4 |
-0.243 |
-0.515 |
| 131 |
258702_at |
AT3G09730
|
unknown |
-0.237 |
-0.071 |