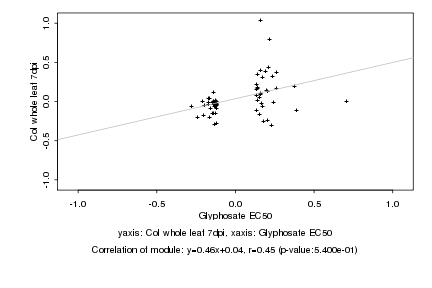
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Glyphosate EC50 |
Log2 signal ratio
Col whole leaf 7dpi |
| 1 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
0.705 |
0.013 |
| 2 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.386 |
-0.111 |
| 3 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.371 |
0.197 |
| 4 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.258 |
0.172 |
| 5 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.256 |
0.379 |
| 6 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.236 |
-0.011 |
| 7 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.231 |
0.324 |
| 8 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.228 |
-0.306 |
| 9 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.216 |
0.795 |
| 10 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.209 |
0.441 |
| 11 |
256924_at |
AT3G29590
|
AT5MAT |
0.202 |
-0.231 |
| 12 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.202 |
0.139 |
| 13 |
263039_at |
AT1G23280
|
[MAK16 protein-related] |
0.196 |
0.151 |
| 14 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.190 |
0.386 |
| 15 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.174 |
-0.247 |
| 16 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.170 |
-0.058 |
| 17 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.166 |
0.319 |
| 18 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.162 |
-0.015 |
| 19 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.159 |
0.093 |
| 20 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.157 |
0.407 |
| 21 |
258396_at |
AT3G15460
|
[Ribosomal RNA processing Brix domain protein] |
0.153 |
0.110 |
| 22 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.153 |
1.047 |
| 23 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
0.148 |
-0.159 |
| 24 |
254455_at |
AT4G21140
|
unknown |
0.147 |
0.055 |
| 25 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.139 |
0.352 |
| 26 |
266139_at |
AT2G28085
|
SAUR42, SMALL AUXIN UPREGULATED RNA 42 |
0.139 |
0.021 |
| 27 |
258202_at |
AT3G13940
|
[DNA binding] |
0.139 |
0.188 |
| 28 |
251538_at |
AT3G58660
|
[Ribosomal protein L1p/L10e family] |
0.134 |
0.174 |
| 29 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
0.133 |
0.230 |
| 30 |
250309_at |
AT5G12220
|
[las1-like family protein] |
0.131 |
0.085 |
| 31 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
0.130 |
0.164 |
| 32 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
0.130 |
-0.109 |
| 33 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.285 |
-0.057 |
| 34 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.244 |
-0.197 |
| 35 |
248073_at |
AT5G55720
|
[Pectin lyase-like superfamily protein] |
-0.214 |
0.009 |
| 36 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
-0.204 |
-0.169 |
| 37 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.202 |
-0.049 |
| 38 |
251050_at |
AT5G02440
|
unknown |
-0.177 |
-0.030 |
| 39 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.177 |
-0.010 |
| 40 |
254954_at |
AT4G10910
|
unknown |
-0.175 |
0.043 |
| 41 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.169 |
-0.201 |
| 42 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.167 |
0.049 |
| 43 |
251976_at |
AT3G53240
|
AtRLP45, receptor like protein 45, RLP45, receptor like protein 45 |
-0.164 |
-0.080 |
| 44 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.150 |
-0.146 |
| 45 |
260515_at |
AT1G51460
|
ABCG13, ATP-binding cassette G13 |
-0.148 |
-0.010 |
| 46 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
-0.145 |
-0.035 |
| 47 |
253573_at |
AT4G31020
|
[alpha/beta-Hydrolases superfamily protein] |
-0.145 |
0.005 |
| 48 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.143 |
-0.146 |
| 49 |
256958_at |
AT3G13430
|
[RING/U-box superfamily protein] |
-0.143 |
0.120 |
| 50 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.142 |
0.011 |
| 51 |
262582_at |
AT1G15410
|
[aspartate-glutamate racemase family] |
-0.139 |
-0.288 |
| 52 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.139 |
-0.062 |
| 53 |
257221_at |
AT3G27920
|
ATGL1, ATMYB0, myb domain protein 0, GL1, GLABRA 1, MYB0, myb domain protein 0 |
-0.137 |
-0.009 |
| 54 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
-0.130 |
-0.063 |
| 55 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.129 |
-0.149 |
| 56 |
260498_at |
AT2G41710
|
[Integrase-type DNA-binding superfamily protein] |
-0.127 |
0.021 |
| 57 |
259174_at |
AT3G03690
|
UNE7, unfertilized embryo sac 7 |
-0.125 |
-0.085 |
| 58 |
251592_at |
AT3G57670
|
NTT, NO TRANSMITTING TRACT, WIP2, WIP domain protein 2 |
-0.124 |
0.003 |
| 59 |
260759_at |
AT1G49180
|
[protein kinase family protein] |
-0.123 |
-0.044 |
| 60 |
248584_at |
AT5G49960
|
unknown |
-0.123 |
-0.025 |
| 61 |
259540_at |
AT1G20640
|
NLP4, NIN-like protein 4 |
-0.123 |
-0.038 |
| 62 |
256640_at |
AT3G32260
|
[Nucleic acid-binding proteins superfamily] |
-0.123 |
-0.026 |
| 63 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.123 |
-0.280 |