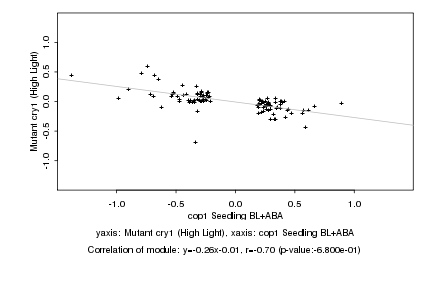
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cop1 Seedling BL+ABA |
Log2 signal ratio
Mutant cry1 (High Light) |
| 1 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.887 |
-0.030 |
| 2 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.664 |
-0.079 |
| 3 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.612 |
-0.149 |
| 4 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.588 |
-0.430 |
| 5 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.565 |
-0.148 |
| 6 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.558 |
-0.200 |
| 7 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.465 |
-0.187 |
| 8 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.441 |
-0.122 |
| 9 |
257456_at |
AT2G18120
|
SRS4, SHI-related sequence 4 |
0.432 |
-0.139 |
| 10 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
0.418 |
-0.260 |
| 11 |
249715_at |
AT5G35760
|
[Beta-galactosidase related protein] |
0.405 |
0.016 |
| 12 |
249747_at |
AT5G24600
|
unknown |
0.396 |
-0.001 |
| 13 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.380 |
0.001 |
| 14 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
0.376 |
-0.035 |
| 15 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
0.375 |
0.006 |
| 16 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.374 |
-0.107 |
| 17 |
256927_at |
AT3G22550
|
unknown |
0.353 |
-0.088 |
| 18 |
249622_at |
AT5G37550
|
unknown |
0.343 |
-0.116 |
| 19 |
267646_at |
AT2G32830
|
PHT1;5, phosphate transporter 1;5, PHT5, PHOSPHATE TRANSPORTER 5 |
0.335 |
0.056 |
| 20 |
258596_at |
AT3G04510
|
LSH2, LIGHT SENSITIVE HYPOCOTYLS 2 |
0.333 |
-0.014 |
| 21 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
0.331 |
-0.297 |
| 22 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
0.326 |
-0.295 |
| 23 |
267070_at |
AT2G41000
|
[Chaperone DnaJ-domain superfamily protein] |
0.316 |
-0.218 |
| 24 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.292 |
-0.079 |
| 25 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.292 |
-0.064 |
| 26 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.290 |
-0.301 |
| 27 |
266337_at |
AT2G32390
|
ATGLR3.5, glutamate receptor 3.5, GLR3.5, glutamate receptor 3.5, GLR6 |
0.290 |
-0.120 |
| 28 |
246682_at |
AT5G33290
|
XGD1, xylogalacturonan deficient 1 |
0.280 |
-0.061 |
| 29 |
259380_at |
AT3G16340
|
ABCG29, ATP-binding cassette G29, AtABCG29, ATPDR1, PLEIOTROPIC DRUG RESISTANCE 1, PDR1, pleiotropic drug resistance 1 |
0.278 |
-0.027 |
| 30 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.276 |
-0.008 |
| 31 |
262634_at |
AT1G06690
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.272 |
-0.147 |
| 32 |
247250_at |
AT5G64630
|
FAS2, FASCIATA 2, MUB3.9, NFB01, NFB1 |
0.265 |
0.054 |
| 33 |
258646_at |
AT3G08040
|
ATFRD3, FRD3, FERRIC REDUCTASE DEFECTIVE 3, MAN1, MANGANESE ACCUMULATOR 1 |
0.259 |
-0.075 |
| 34 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.257 |
-0.119 |
| 35 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.256 |
-0.037 |
| 36 |
249811_at |
AT5G23760
|
[Copper transport protein family] |
0.253 |
-0.067 |
| 37 |
257466_at |
AT1G62840
|
unknown |
0.246 |
0.009 |
| 38 |
260039_at |
AT1G68795
|
CLE12, CLAVATA3/ESR-RELATED 12 |
0.240 |
-0.061 |
| 39 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
0.233 |
-0.166 |
| 40 |
266015_at |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
0.230 |
-0.085 |
| 41 |
248070_at |
AT5G55660
|
[DEK domain-containing chromatin associated protein] |
0.230 |
-0.004 |
| 42 |
257666_at |
AT3G20270
|
[lipid-binding serum glycoprotein family protein] |
0.224 |
0.003 |
| 43 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.217 |
-0.172 |
| 44 |
258738_at |
AT3G05750
|
TRM6, TON1 Recruiting Motif 6 |
0.214 |
-0.028 |
| 45 |
248132_at |
AT5G54840
|
ATSGP1, SGP1 |
0.212 |
-0.022 |
| 46 |
256714_at |
AT2G34080
|
[Cysteine proteinases superfamily protein] |
0.207 |
0.022 |
| 47 |
251672_at |
AT3G57230
|
AGL16, AGAMOUS-like 16 |
0.202 |
-0.041 |
| 48 |
261551_at |
AT1G63440
|
HMA5, heavy metal atpase 5 |
0.197 |
0.040 |
| 49 |
253977_at |
AT4G26630
|
[DEK domain-containing chromatin associated protein] |
0.197 |
0.030 |
| 50 |
248239_at |
AT5G53920
|
[ribosomal protein L11 methyltransferase-related] |
0.194 |
-0.038 |
| 51 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.191 |
-0.052 |
| 52 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.189 |
-0.096 |
| 53 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
0.187 |
-0.201 |
| 54 |
265959_at |
AT2G37240
|
[Thioredoxin superfamily protein] |
0.186 |
-0.037 |
| 55 |
247060_at |
AT5G66760
|
SDH1-1, succinate dehydrogenase 1-1 |
0.186 |
-0.060 |
| 56 |
250084_at |
AT5G17240
|
SDG40, SET domain group 40 |
0.184 |
-0.067 |
| 57 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-1.386 |
0.444 |
| 58 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.986 |
0.066 |
| 59 |
247474_at |
AT5G62280
|
unknown |
-0.908 |
0.218 |
| 60 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
-0.796 |
0.478 |
| 61 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.746 |
0.596 |
| 62 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
-0.721 |
0.131 |
| 63 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.697 |
0.095 |
| 64 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.686 |
0.449 |
| 65 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.652 |
0.384 |
| 66 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
-0.626 |
-0.095 |
| 67 |
259996_at |
AT1G67910
|
unknown |
-0.545 |
0.100 |
| 68 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.535 |
0.120 |
| 69 |
254193_at |
AT4G23870
|
unknown |
-0.527 |
0.159 |
| 70 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.525 |
0.150 |
| 71 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.495 |
0.090 |
| 72 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.479 |
0.003 |
| 73 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-0.478 |
0.038 |
| 74 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.454 |
0.279 |
| 75 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.443 |
0.116 |
| 76 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.420 |
0.130 |
| 77 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
-0.403 |
0.032 |
| 78 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.392 |
-0.011 |
| 79 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
-0.379 |
0.025 |
| 80 |
264590_at |
AT2G17710
|
unknown |
-0.363 |
-0.004 |
| 81 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.360 |
0.002 |
| 82 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.351 |
-0.002 |
| 83 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.345 |
0.023 |
| 84 |
256452_at |
AT1G75240
|
AtHB33, homeobox protein 33, HB33, homeobox protein 33, ZHD5, zinc-finger homeodomain 5 |
-0.344 |
-0.683 |
| 85 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.335 |
0.254 |
| 86 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.327 |
0.134 |
| 87 |
245141_at |
AT2G45400
|
BEN1, BRI1-5 ENHANCED 1 |
-0.326 |
0.045 |
| 88 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.326 |
-0.163 |
| 89 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
-0.322 |
0.138 |
| 90 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
-0.307 |
0.033 |
| 91 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
-0.301 |
0.119 |
| 92 |
245574_at |
AT4G14750
|
IQD19, IQ-domain 19 |
-0.300 |
0.089 |
| 93 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
-0.298 |
0.006 |
| 94 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.293 |
0.175 |
| 95 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.292 |
0.015 |
| 96 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
-0.289 |
0.160 |
| 97 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
-0.281 |
0.033 |
| 98 |
253218_at |
AT4G34980
|
SLP2, subtilisin-like serine protease 2 |
-0.280 |
0.096 |
| 99 |
263218_at |
AT1G30570
|
HERK2, hercules receptor kinase 2 |
-0.273 |
0.106 |
| 100 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
-0.271 |
0.051 |
| 101 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.270 |
0.075 |
| 102 |
261949_at |
AT1G64670
|
BDG1, BODYGUARD1, CED1, 9-CIS EPOXYCAROTENOID DIOXYGENASE DEFECTIVE 1 |
-0.269 |
0.002 |
| 103 |
246565_at |
AT5G15530
|
BCCP2, biotin carboxyl carrier protein 2, CAC1-B |
-0.256 |
0.040 |
| 104 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.250 |
0.103 |
| 105 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
-0.250 |
0.020 |
| 106 |
248177_at |
AT5G54630
|
[zinc finger protein-related] |
-0.243 |
0.153 |
| 107 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.239 |
0.140 |
| 108 |
246483_at |
AT5G16000
|
AtNIK1, NIK1, NSP-interacting kinase 1 |
-0.235 |
0.071 |
| 109 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.227 |
0.166 |
| 110 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.225 |
0.089 |
| 111 |
260668_at |
AT1G19530
|
unknown |
-0.216 |
0.002 |