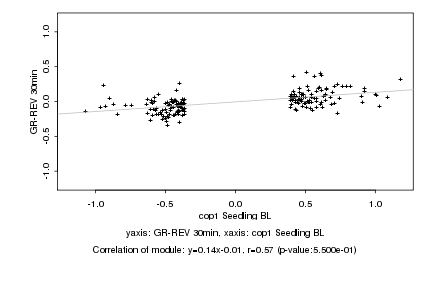
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cop1 Seedling BL |
Log2 signal ratio
GR-REV 30min |
| 1 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
1.177 |
0.326 |
| 2 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
1.086 |
0.060 |
| 3 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
1.024 |
-0.070 |
| 4 |
245799_at |
AT1G45616
|
AtRLP6, receptor like protein 6, RLP6, receptor like protein 6 |
1.006 |
0.100 |
| 5 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.994 |
0.104 |
| 6 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.920 |
0.147 |
| 7 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.920 |
0.198 |
| 8 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.907 |
-0.004 |
| 9 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
0.896 |
0.082 |
| 10 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.820 |
0.224 |
| 11 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.793 |
0.223 |
| 12 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.762 |
0.228 |
| 13 |
267548_at |
AT2G32660
|
AtRLP22, receptor like protein 22, RLP22, receptor like protein 22 |
0.737 |
0.044 |
| 14 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.728 |
0.246 |
| 15 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.723 |
-0.164 |
| 16 |
246072_at |
AT5G20240
|
PI, PISTILLATA |
0.705 |
-0.027 |
| 17 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.705 |
0.219 |
| 18 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.688 |
0.140 |
| 19 |
258327_at |
AT3G22640
|
PAP85 |
0.686 |
-0.041 |
| 20 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.673 |
0.061 |
| 21 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.649 |
0.175 |
| 22 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.647 |
0.194 |
| 23 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.641 |
0.097 |
| 24 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.639 |
0.015 |
| 25 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.637 |
0.107 |
| 26 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.623 |
0.074 |
| 27 |
259150_at |
AT3G10320
|
[Glycosyltransferase family 61 protein] |
0.619 |
-0.085 |
| 28 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.615 |
0.159 |
| 29 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.613 |
0.376 |
| 30 |
246949_at |
AT5G25140
|
CYP71B13, cytochrome P450, family 71, subfamily B, polypeptide 13 |
0.610 |
-0.014 |
| 31 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
0.607 |
-0.035 |
| 32 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.604 |
0.404 |
| 33 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.597 |
0.214 |
| 34 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.592 |
0.190 |
| 35 |
250875_at |
AT5G04020
|
[calmodulin binding] |
0.579 |
0.046 |
| 36 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.576 |
0.154 |
| 37 |
255306_at |
AT4G04740
|
ATCPK23, CPK23, calcium-dependent protein kinase 23 |
0.575 |
0.002 |
| 38 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
0.575 |
-0.080 |
| 39 |
265660_at |
AT2G25470
|
AtRLP21, receptor like protein 21, RLP21, receptor like protein 21 |
0.560 |
0.366 |
| 40 |
260031_at |
AT1G68790
|
CRWN3, CROWDED NUCLEI 3, LINC3, LITTLE NUCLEI3 |
0.558 |
0.049 |
| 41 |
249622_at |
AT5G37550
|
unknown |
0.548 |
-0.126 |
| 42 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.542 |
-0.003 |
| 43 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
0.541 |
0.103 |
| 44 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.537 |
0.060 |
| 45 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.534 |
-0.091 |
| 46 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.529 |
-0.002 |
| 47 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.525 |
0.019 |
| 48 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.522 |
0.172 |
| 49 |
250660_at |
AT5G07060
|
MAC5C, MOS4-associated complex subunit 5C |
0.517 |
-0.013 |
| 50 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.511 |
0.012 |
| 51 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.509 |
0.228 |
| 52 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.506 |
-0.073 |
| 53 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.503 |
0.429 |
| 54 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.500 |
-0.016 |
| 55 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.494 |
0.070 |
| 56 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
0.490 |
-0.017 |
| 57 |
248228_at |
AT5G53800
|
unknown |
0.484 |
0.065 |
| 58 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.482 |
0.105 |
| 59 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.476 |
0.112 |
| 60 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
0.475 |
-0.069 |
| 61 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.471 |
0.113 |
| 62 |
263569_at |
AT2G27170
|
SMC3, STRUCTURAL MAINTENANCE OF CHROMOSOMES 3, TTN7, TITAN7 |
0.471 |
0.014 |
| 63 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
0.470 |
0.065 |
| 64 |
265678_at |
AT2G31970
|
ATRAD50, RAD50 |
0.467 |
0.042 |
| 65 |
256659_at |
AT3G12020
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.464 |
0.015 |
| 66 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.457 |
0.071 |
| 67 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
0.456 |
0.135 |
| 68 |
252611_at |
AT3G45130
|
LAS1, lanosterol synthase 1 |
0.452 |
0.192 |
| 69 |
256544_at |
AT1G42560
|
ATMLO9, ARABIDOPSIS THALIANA MILDEW RESISTANCE LOCUS O 9, MLO9, MILDEW RESISTANCE LOCUS O 9 |
0.445 |
0.030 |
| 70 |
249409_at |
AT5G40340
|
[Tudor/PWWP/MBT superfamily protein] |
0.444 |
-0.025 |
| 71 |
256096_at |
AT1G13650
|
unknown |
0.441 |
-0.011 |
| 72 |
260296_at |
AT1G63750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.435 |
0.082 |
| 73 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.430 |
-0.126 |
| 74 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
0.428 |
-0.005 |
| 75 |
248344_at |
AT5G52280
|
[Myosin heavy chain-related protein] |
0.426 |
0.018 |
| 76 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
0.423 |
-0.110 |
| 77 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
0.422 |
0.114 |
| 78 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.420 |
0.067 |
| 79 |
263380_at |
AT2G40200
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.414 |
0.369 |
| 80 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.410 |
0.144 |
| 81 |
256425_at |
AT1G33560
|
ADR1, ACTIVATED DISEASE RESISTANCE 1 |
0.409 |
0.070 |
| 82 |
245702_at |
AT5G04220
|
ATSYTC, NTMC2T1.3, NTMC2TYPE1.3, SYT3, synaptotagmin 3, SYTC |
0.409 |
0.049 |
| 83 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
0.406 |
0.051 |
| 84 |
257073_at |
AT3G19650
|
[cyclin-related] |
0.405 |
0.027 |
| 85 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.405 |
0.065 |
| 86 |
265196_at |
AT2G36740
|
ATSWC2, SWC2 |
0.400 |
0.049 |
| 87 |
258833_at |
AT3G07274
|
|
0.399 |
0.104 |
| 88 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.397 |
0.078 |
| 89 |
247835_at |
AT5G57910
|
unknown |
0.396 |
-0.037 |
| 90 |
255836_at |
AT2G33440
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.394 |
0.051 |
| 91 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.391 |
-0.085 |
| 92 |
253977_at |
AT4G26630
|
[DEK domain-containing chromatin associated protein] |
0.390 |
0.020 |
| 93 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
0.388 |
0.077 |
| 94 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-1.072 |
-0.138 |
| 95 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.971 |
-0.075 |
| 96 |
262373_at |
AT1G73120
|
unknown |
-0.949 |
0.241 |
| 97 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.935 |
-0.069 |
| 98 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.902 |
0.056 |
| 99 |
265246_at |
AT2G43050
|
ATPMEPCRD |
-0.877 |
-0.038 |
| 100 |
253155_at |
AT4G35720
|
unknown |
-0.850 |
-0.184 |
| 101 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.791 |
-0.046 |
| 102 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.746 |
-0.049 |
| 103 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.636 |
-0.036 |
| 104 |
258196_at |
AT3G13980
|
unknown |
-0.632 |
0.042 |
| 105 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.630 |
-0.160 |
| 106 |
262446_at |
AT1G49310
|
unknown |
-0.614 |
-0.106 |
| 107 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.609 |
-0.269 |
| 108 |
259897_at |
AT1G71380
|
ATCEL3, cellulase 3, ATGH9B3, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B3, CEL3, cellulase 3 |
-0.600 |
0.022 |
| 109 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.600 |
-0.012 |
| 110 |
247091_at |
AT5G66390
|
PRX72, PEROXIDASE 72 |
-0.596 |
-0.199 |
| 111 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.595 |
-0.027 |
| 112 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.592 |
-0.106 |
| 113 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.580 |
0.066 |
| 114 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.579 |
0.009 |
| 115 |
259276_at |
AT3G01190
|
[Peroxidase superfamily protein] |
-0.575 |
-0.122 |
| 116 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.569 |
-0.186 |
| 117 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.569 |
-0.087 |
| 118 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.555 |
0.114 |
| 119 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.553 |
-0.183 |
| 120 |
265031_at |
AT1G61590
|
[Protein kinase superfamily protein] |
-0.541 |
-0.152 |
| 121 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
-0.534 |
-0.179 |
| 122 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.523 |
-0.122 |
| 123 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.522 |
-0.257 |
| 124 |
263437_at |
AT2G28670
|
ESB1, ENHANCED SUBERIN 1 |
-0.515 |
-0.203 |
| 125 |
261745_at |
AT1G08500
|
AtENODL18, ENODL18, early nodulin-like protein 18 |
-0.506 |
-0.106 |
| 126 |
246142_at |
AT5G19970
|
unknown |
-0.506 |
-0.026 |
| 127 |
254718_at |
AT4G13580
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.500 |
-0.242 |
| 128 |
251358_at |
AT3G61160
|
[Protein kinase superfamily protein] |
-0.496 |
-0.156 |
| 129 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.495 |
-0.285 |
| 130 |
261641_at |
AT1G27670
|
unknown |
-0.490 |
-0.006 |
| 131 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
-0.487 |
-0.333 |
| 132 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
-0.486 |
-0.203 |
| 133 |
263002_at |
AT1G54200
|
unknown |
-0.484 |
-0.048 |
| 134 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.479 |
-0.227 |
| 135 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.478 |
-0.178 |
| 136 |
246565_at |
AT5G15530
|
BCCP2, biotin carboxyl carrier protein 2, CAC1-B |
-0.472 |
-0.033 |
| 137 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.471 |
-0.121 |
| 138 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.467 |
-0.091 |
| 139 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.461 |
-0.070 |
| 140 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.460 |
0.031 |
| 141 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.459 |
0.004 |
| 142 |
267006_at |
AT2G34190
|
[Xanthine/uracil permease family protein] |
-0.454 |
-0.011 |
| 143 |
253580_at |
AT4G30400
|
[RING/U-box superfamily protein] |
-0.445 |
-0.078 |
| 144 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.445 |
-0.007 |
| 145 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
-0.444 |
-0.190 |
| 146 |
259291_at |
AT3G11550
|
CASP2, Casparian strip membrane domain protein 2 |
-0.441 |
-0.197 |
| 147 |
262388_at |
AT1G49320
|
unknown |
-0.441 |
0.023 |
| 148 |
265645_at |
AT2G27370
|
CASP3, Casparian strip membrane domain protein 3 |
-0.430 |
-0.179 |
| 149 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.429 |
0.027 |
| 150 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.424 |
0.169 |
| 151 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.424 |
-0.039 |
| 152 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.423 |
0.005 |
| 153 |
246425_at |
AT5G17420
|
ATCESA7, CESA7, CELLULOSE SYNTHASE CATALYTIC SUBUNIT 7, IRX3, IRREGULAR XYLEM 3, MUR10, MURUS 10 |
-0.418 |
-0.131 |
| 154 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
-0.418 |
-0.071 |
| 155 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.417 |
-0.148 |
| 156 |
252172_at |
AT3G50640
|
unknown |
-0.414 |
-0.134 |
| 157 |
250982_at |
AT5G03150
|
JKD, JACKDAW |
-0.413 |
-0.071 |
| 158 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.413 |
-0.181 |
| 159 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.411 |
-0.127 |
| 160 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.409 |
-0.023 |
| 161 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
-0.407 |
-0.053 |
| 162 |
252129_at |
AT3G50890
|
AtHB28, homeobox protein 28, HB28, homeobox protein 28, ZHD7, ZINC FINGER HOMEODOMAIN 7 |
-0.405 |
-0.044 |
| 163 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-0.401 |
-0.300 |
| 164 |
266783_at |
AT2G29130
|
ATLAC2, LAC2, laccase 2 |
-0.401 |
-0.141 |
| 165 |
259680_at |
AT1G77690
|
LAX3, like AUX1 3 |
-0.400 |
-0.056 |
| 166 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
-0.400 |
0.259 |
| 167 |
261893_at |
AT1G80690
|
[PPPDE putative thiol peptidase family protein] |
-0.399 |
-0.072 |
| 168 |
247522_at |
AT5G61340
|
unknown |
-0.396 |
-0.024 |
| 169 |
260363_at |
AT1G70550
|
unknown |
-0.388 |
-0.004 |
| 170 |
246135_at |
AT5G20885
|
[RING/U-box superfamily protein] |
-0.387 |
-0.044 |
| 171 |
265250_at |
AT2G01950
|
BRL2, BRI1-like 2, VH1, VASCULAR HIGHWAY 1 |
-0.385 |
0.036 |
| 172 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
-0.385 |
-0.194 |
| 173 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.384 |
0.033 |
| 174 |
263373_at |
AT2G20515
|
unknown |
-0.384 |
-0.036 |
| 175 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
-0.382 |
-0.127 |
| 176 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
-0.379 |
-0.093 |
| 177 |
266688_at |
AT2G19660
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.375 |
0.023 |
| 178 |
258445_at |
AT3G22400
|
ATLOX5, Arabidopsis thaliana lipoxygenase 5, LOX5 |
-0.373 |
-0.107 |
| 179 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.373 |
-0.021 |
| 180 |
246908_at |
AT5G25610
|
ATRD22, RD22, RESPONSIVE TO DESSICATION 22 |
-0.372 |
-0.031 |
| 181 |
256621_at |
AT3G24450
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.371 |
0.038 |
| 182 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
-0.370 |
-0.177 |
| 183 |
248646_at |
AT5G49100
|
unknown |
-0.369 |
-0.017 |
| 184 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.369 |
-0.086 |
| 185 |
249885_at |
AT5G22940
|
F8H, FRA8 homolog |
-0.366 |
-0.130 |