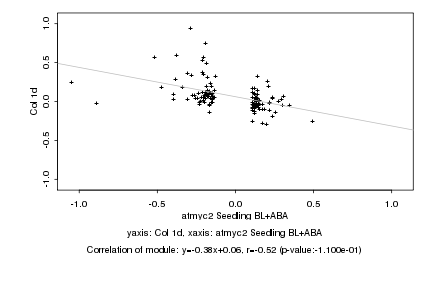
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
atmyc2 Seedling BL+ABA |
Log2 signal ratio
Col 1d |
| 1 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
0.489 |
-0.250 |
| 2 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
0.340 |
-0.046 |
| 3 |
251458_at |
AT3G60170
|
[copia-like retrotransposon family, has a 7.6e-229 P-value blast match to gb|AAO73527.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.304 |
0.068 |
| 4 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
0.299 |
-0.041 |
| 5 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
0.293 |
0.030 |
| 6 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
0.275 |
0.006 |
| 7 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
0.256 |
-0.136 |
| 8 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
0.233 |
-0.183 |
| 9 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
0.232 |
0.058 |
| 10 |
257853_at |
AT3G12960
|
SMP1, seed maturation protein 1 |
0.231 |
0.050 |
| 11 |
253977_at |
AT4G26630
|
[DEK domain-containing chromatin associated protein] |
0.214 |
-0.007 |
| 12 |
258626_at |
AT3G04450
|
[Homeodomain-like superfamily protein] |
0.214 |
-0.114 |
| 13 |
248070_at |
AT5G55660
|
[DEK domain-containing chromatin associated protein] |
0.212 |
-0.021 |
| 14 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.206 |
0.202 |
| 15 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
0.202 |
0.261 |
| 16 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
0.196 |
-0.283 |
| 17 |
264454_at |
AT1G10320
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.183 |
-0.095 |
| 18 |
248291_at |
AT5G53020
|
[Ribonuclease P protein subunit P38-related] |
0.169 |
-0.273 |
| 19 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
0.167 |
-0.029 |
| 20 |
245618_at |
AT4G14510
|
ATCFM3B, CFM3B, CRM family member 3B |
0.167 |
-0.095 |
| 21 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
0.153 |
0.024 |
| 22 |
263007_at |
AT1G54260
|
[winged-helix DNA-binding transcription factor family protein] |
0.150 |
-0.099 |
| 23 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
0.148 |
-0.037 |
| 24 |
247004_at |
AT5G67570
|
DG1, DELAYED GREENING 1, EMB1408, embryo defective 1408, EMB246, EMBRYO DEFECTIVE 246 |
0.147 |
-0.070 |
| 25 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.146 |
-0.032 |
| 26 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.146 |
-0.041 |
| 27 |
266727_at |
AT2G03150
|
emb1579, embryo defective 1579 |
0.144 |
-0.013 |
| 28 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
0.144 |
0.009 |
| 29 |
251829_at |
AT3G55020
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.141 |
0.045 |
| 30 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
0.141 |
0.323 |
| 31 |
251466_at |
AT3G59340
|
unknown |
0.139 |
0.091 |
| 32 |
245725_at |
AT1G73370
|
ATSUS6, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 6, SUS6, sucrose synthase 6 |
0.135 |
0.149 |
| 33 |
257134_at |
AT3G12870
|
unknown |
0.134 |
0.043 |
| 34 |
263696_at |
AT1G31230
|
AK-HSDH, ASPARTATE KINASE-HOMOSERINE DEHYDROGENASE, AK-HSDH I, aspartate kinase-homoserine dehydrogenase i |
0.133 |
-0.033 |
| 35 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
0.133 |
0.010 |
| 36 |
247883_at |
AT5G57790
|
unknown |
0.133 |
0.024 |
| 37 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
0.129 |
0.055 |
| 38 |
248938_at |
AT5G45780
|
[Leucine-rich repeat protein kinase family protein] |
0.129 |
-0.055 |
| 39 |
266754_at |
AT2G46980
|
ASY3, ASYNAPTIC 3, AtASY3, Arabidopsis thaliana ASYNAPTIC 3 |
0.128 |
0.024 |
| 40 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
0.127 |
-0.026 |
| 41 |
265957_at |
AT2G37300
|
ABCI16, ATP-binding cassette I16 |
0.125 |
0.043 |
| 42 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.124 |
-0.054 |
| 43 |
246898_at |
AT5G25580
|
unknown |
0.122 |
0.064 |
| 44 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.121 |
0.174 |
| 45 |
252373_at |
AT3G48090
|
ATEDS1, EDS1, enhanced disease susceptibility 1 |
0.120 |
-0.121 |
| 46 |
255904_at |
AT1G17860
|
[Kunitz family trypsin and protease inhibitor protein] |
0.120 |
0.025 |
| 47 |
247046_at |
AT5G66540
|
unknown |
0.119 |
0.097 |
| 48 |
249362_at |
AT5G40550
|
AtSGF29b, SGF29b, SaGa associated Factor 29 b |
0.119 |
-0.151 |
| 49 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
0.118 |
0.110 |
| 50 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
0.118 |
-0.035 |
| 51 |
263714_at |
AT2G20610
|
ALF1, ABERRANT LATERAL ROOT FORMATION 1, HLS3, HOOKLESS 3, RTY, ROOTY, RTY1, ROOTY 1, SUR1, SUPERROOT 1 |
0.118 |
-0.065 |
| 52 |
262006_at |
AT1G64570
|
DUO3, DUO POLLEN 3 |
0.117 |
-0.078 |
| 53 |
263202_at |
AT1G05630
|
5PTASE13, inositol-polyphosphate 5-phosphatase 13, AT5PTASE13, Arabidopsis thaliana inositol-polyphosphate 5-phosphatase 13 |
0.114 |
-0.038 |
| 54 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
0.114 |
-0.051 |
| 55 |
262847_at |
AT1G14840
|
ATMAP70-4, microtubule-associated proteins 70-4, MAP70-4, microtubule-associated proteins 70-4 |
0.109 |
-0.036 |
| 56 |
255480_at |
AT4G02485
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.108 |
-0.106 |
| 57 |
252432_at |
AT3G47680
|
[DNA binding] |
0.107 |
-0.087 |
| 58 |
250501_at |
AT5G09640
|
SCPL19, serine carboxypeptidase-like 19, SNG2, SINAPOYLGLUCOSE ACCUMULATOR 2 |
0.107 |
0.121 |
| 59 |
257547_at |
AT3G13000
|
unknown |
0.106 |
-0.065 |
| 60 |
249231_at |
AT5G42030
|
ABIL4, ABL interactor-like protein 4 |
0.105 |
-0.005 |
| 61 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
0.104 |
0.177 |
| 62 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
0.104 |
0.053 |
| 63 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
0.103 |
-0.245 |
| 64 |
256527_at |
AT1G66100
|
[Plant thionin] |
-1.054 |
0.253 |
| 65 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
-0.892 |
-0.022 |
| 66 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
-0.519 |
0.573 |
| 67 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
-0.477 |
0.192 |
| 68 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
-0.398 |
0.098 |
| 69 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.397 |
0.034 |
| 70 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.389 |
0.286 |
| 71 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
-0.379 |
0.596 |
| 72 |
266658_at |
AT2G25735
|
unknown |
-0.344 |
0.193 |
| 73 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
-0.310 |
0.034 |
| 74 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.307 |
0.365 |
| 75 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.290 |
0.943 |
| 76 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.286 |
0.344 |
| 77 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.275 |
0.089 |
| 78 |
263851_at |
AT2G04460
|
unknown |
-0.268 |
0.086 |
| 79 |
262857_at |
AT1G14930
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.261 |
0.041 |
| 80 |
259935_at |
AT1G71250
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.247 |
0.048 |
| 81 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
-0.239 |
0.106 |
| 82 |
267177_at |
AT2G37580
|
[RING/U-box superfamily protein] |
-0.236 |
-0.031 |
| 83 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
-0.236 |
-0.012 |
| 84 |
261215_at |
AT1G32970
|
[Subtilisin-like serine endopeptidase family protein] |
-0.226 |
0.007 |
| 85 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
-0.220 |
0.061 |
| 86 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
-0.216 |
0.536 |
| 87 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
-0.215 |
0.120 |
| 88 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
-0.213 |
0.382 |
| 89 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
-0.210 |
0.063 |
| 90 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
-0.209 |
0.348 |
| 91 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
-0.208 |
0.020 |
| 92 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.208 |
0.577 |
| 93 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
-0.204 |
0.089 |
| 94 |
247953_at |
AT5G57260
|
CYP71B10, cytochrome P450, family 71, subfamily B, polypeptide 10 |
-0.202 |
-0.005 |
| 95 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
-0.198 |
0.757 |
| 96 |
257321_at |
ATMG01130
|
ORF106F |
-0.197 |
0.128 |
| 97 |
244901_at |
ATMG00640
|
ORF25 |
-0.197 |
0.092 |
| 98 |
256712_at |
AT2G34020
|
[Calcium-binding EF-hand family protein] |
-0.197 |
0.048 |
| 99 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.189 |
0.112 |
| 100 |
253827_at |
AT4G28085
|
unknown |
-0.189 |
0.497 |
| 101 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.186 |
0.200 |
| 102 |
246270_at |
AT4G36500
|
unknown |
-0.185 |
0.151 |
| 103 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.185 |
0.066 |
| 104 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.181 |
0.081 |
| 105 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.180 |
0.311 |
| 106 |
249252_at |
AT5G42010
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.177 |
0.098 |
| 107 |
258610_at |
AT3G02875
|
ILR1, IAA-LEUCINE RESISTANT 1 |
-0.175 |
0.067 |
| 108 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
-0.170 |
0.137 |
| 109 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
-0.170 |
-0.031 |
| 110 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.170 |
-0.047 |
| 111 |
257369_at |
AT2G35550
|
ATBPC7, BASIC PENTACYSTEINE 7, BBR, BPC7, basic pentacysteine 7 |
-0.167 |
-0.137 |
| 112 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
-0.164 |
0.050 |
| 113 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
-0.164 |
0.242 |
| 114 |
245041_at |
AT2G26530
|
AR781 |
-0.163 |
0.107 |
| 115 |
244945_at |
ATMG00110
|
ABCI2, ATP-binding cassette I2, CCB206, cytochrome C biogenesis 206 |
-0.159 |
-0.002 |
| 116 |
253047_at |
AT4G37295
|
unknown |
-0.156 |
0.196 |
| 117 |
261350_at |
AT1G79770
|
unknown |
-0.156 |
0.101 |
| 118 |
245000_at |
ATCG00210
|
YCF6 |
-0.153 |
0.115 |
| 119 |
257322_at |
ATMG01180
|
ORF111B |
-0.153 |
-0.011 |
| 120 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.153 |
0.073 |
| 121 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
-0.150 |
0.034 |
| 122 |
257274_at |
AT3G14510
|
[Polyprenyl synthetase family protein] |
-0.145 |
0.060 |
| 123 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
-0.137 |
0.053 |
| 124 |
265057_at |
AT1G52140
|
unknown |
-0.135 |
0.145 |
| 125 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
-0.133 |
0.327 |