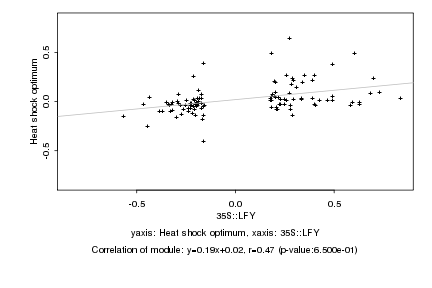
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S::LFY |
Log2 signal ratio
Heat shock optimum |
| 1 |
254452_at |
AT4G21100
|
DDB1B, damaged DNA binding protein 1B |
0.834 |
0.040 |
| 2 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.727 |
0.100 |
| 3 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.698 |
0.242 |
| 4 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.680 |
0.091 |
| 5 |
246531_at |
AT5G15800
|
AGL2, AGAMOUS-like 2, SEP1, SEPALLATA1 |
0.628 |
-0.030 |
| 6 |
247490_at |
AT5G61850
|
LFY, LEAFY, LFY3, LEAFY 3 |
0.625 |
-0.004 |
| 7 |
261684_at |
AT1G47400
|
unknown |
0.600 |
0.496 |
| 8 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.591 |
-0.001 |
| 9 |
247610_at |
AT5G60630
|
unknown |
0.579 |
-0.038 |
| 10 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.491 |
0.056 |
| 11 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.490 |
0.019 |
| 12 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.487 |
0.380 |
| 13 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.462 |
0.013 |
| 14 |
251293_at |
AT3G61930
|
unknown |
0.424 |
0.012 |
| 15 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
0.402 |
-0.034 |
| 16 |
264635_at |
AT1G65500
|
unknown |
0.398 |
0.274 |
| 17 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.397 |
-0.027 |
| 18 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.390 |
0.221 |
| 19 |
251363_at |
AT3G61250
|
AtMYB17, myb domain protein 17, LMI2, LATE MERISTEM IDENTITY2, MYB17, myb domain protein 17 |
0.386 |
0.039 |
| 20 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
0.348 |
0.266 |
| 21 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.337 |
0.198 |
| 22 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.334 |
0.026 |
| 23 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
0.332 |
0.039 |
| 24 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
0.306 |
0.143 |
| 25 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.294 |
0.029 |
| 26 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.292 |
0.223 |
| 27 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
0.287 |
-0.136 |
| 28 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
0.286 |
0.239 |
| 29 |
250828_at |
AT5G05250
|
unknown |
0.282 |
0.181 |
| 30 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.275 |
-0.080 |
| 31 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
0.274 |
-0.033 |
| 32 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.272 |
0.085 |
| 33 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.269 |
0.650 |
| 34 |
248864_at |
AT5G46760
|
MYC3 |
0.258 |
0.016 |
| 35 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.258 |
0.268 |
| 36 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.244 |
0.021 |
| 37 |
248868_at |
AT5G46780
|
[VQ motif-containing protein] |
0.244 |
-0.027 |
| 38 |
245296_at |
AT4G16370
|
oligopeptide transporter |
0.228 |
0.024 |
| 39 |
257421_at |
AT1G12030
|
unknown |
0.224 |
-0.028 |
| 40 |
247001_at |
AT5G67330
|
ATNRAMP4, ARABIDOPSIS THALIANA NATURAL RESISTANCE ASSOCIATED MACROPHAGE PROTEIN 4, NRAMP4, natural resistance associated macrophage protein 4 |
0.223 |
-0.024 |
| 41 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.214 |
0.048 |
| 42 |
259999_at |
AT1G68080
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.211 |
-0.075 |
| 43 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
0.206 |
-0.057 |
| 44 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.204 |
-0.077 |
| 45 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.202 |
0.200 |
| 46 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.198 |
0.093 |
| 47 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
0.198 |
0.051 |
| 48 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.195 |
0.210 |
| 49 |
257062_at |
AT3G18290
|
BTS, BRUTUS, EMB2454, embryo defective 2454 |
0.193 |
0.061 |
| 50 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
0.186 |
0.080 |
| 51 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
0.182 |
0.058 |
| 52 |
255733_at |
AT1G25400
|
unknown |
0.182 |
-0.054 |
| 53 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
0.181 |
0.015 |
| 54 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.180 |
0.493 |
| 55 |
251373_at |
AT3G60530
|
GATA4, GATA transcription factor 4 |
0.173 |
0.040 |
| 56 |
262529_at |
AT1G17250
|
AtRLP3, receptor like protein 3, RFO2, RESISTANCE TO FUSARIUM OXYSPORUM 2, RLP3, receptor like protein 3 |
0.173 |
0.013 |
| 57 |
256096_at |
AT1G13650
|
unknown |
-0.570 |
-0.151 |
| 58 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.470 |
-0.022 |
| 59 |
247878_at |
AT5G57760
|
unknown |
-0.446 |
-0.249 |
| 60 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.437 |
0.051 |
| 61 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.385 |
-0.093 |
| 62 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.373 |
-0.092 |
| 63 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.352 |
-0.001 |
| 64 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
-0.341 |
-0.021 |
| 65 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
-0.338 |
-0.039 |
| 66 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
-0.329 |
-0.100 |
| 67 |
266403_at |
AT2G38600
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.324 |
-0.025 |
| 68 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.319 |
-0.007 |
| 69 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.319 |
-0.089 |
| 70 |
252073_at |
AT3G51750
|
unknown |
-0.299 |
-0.155 |
| 71 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.296 |
0.003 |
| 72 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.291 |
-0.018 |
| 73 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
-0.290 |
0.080 |
| 74 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.281 |
-0.036 |
| 75 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
-0.274 |
-0.130 |
| 76 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.267 |
-0.072 |
| 77 |
262080_at |
AT1G59650
|
CW14 |
-0.254 |
-0.037 |
| 78 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.252 |
0.014 |
| 79 |
256891_at |
AT3G19030
|
unknown |
-0.239 |
-0.101 |
| 80 |
252882_at |
AT4G39675
|
unknown |
-0.238 |
-0.064 |
| 81 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.230 |
-0.034 |
| 82 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.229 |
-0.020 |
| 83 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.229 |
-0.068 |
| 84 |
253515_at |
AT4G31320
|
SAUR37, SMALL AUXIN UPREGULATED RNA 37 |
-0.227 |
-0.036 |
| 85 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
-0.225 |
-0.019 |
| 86 |
261448_at |
AT1G21140
|
[Vacuolar iron transporter (VIT) family protein] |
-0.219 |
-0.119 |
| 87 |
260668_at |
AT1G19530
|
unknown |
-0.217 |
0.025 |
| 88 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.216 |
0.257 |
| 89 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.214 |
-0.046 |
| 90 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
-0.209 |
-0.078 |
| 91 |
254057_at |
AT4G25170
|
[Uncharacterised conserved protein (UCP012943)] |
-0.209 |
0.020 |
| 92 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
-0.206 |
-0.021 |
| 93 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.204 |
-0.139 |
| 94 |
262533_at |
AT1G17090
|
unknown |
-0.201 |
-0.007 |
| 95 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-0.198 |
-0.042 |
| 96 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
-0.196 |
-0.036 |
| 97 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.194 |
0.038 |
| 98 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
-0.190 |
-0.011 |
| 99 |
250475_at |
AT5G10180
|
AST68, ARABIDOPSIS SULFATE TRANSPORTER 68, SULTR2;1, sulfate transporter 2;1 |
-0.189 |
0.009 |
| 100 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
-0.188 |
0.119 |
| 101 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
-0.186 |
0.033 |
| 102 |
254240_at |
AT4G23496
|
SP1L5, SPIRAL1-like5 |
-0.176 |
0.037 |
| 103 |
262831_at |
AT1G14730
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.173 |
-0.011 |
| 104 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.173 |
0.075 |
| 105 |
255691_at |
AT4G00370
|
ANTR2, PHT4;4, anion transporter 2 |
-0.173 |
-0.062 |
| 106 |
246142_at |
AT5G19970
|
unknown |
-0.170 |
-0.179 |
| 107 |
258184_at |
AT3G21510
|
AHP1, histidine-containing phosphotransmitter 1 |
-0.166 |
-0.132 |
| 108 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-0.163 |
-0.049 |
| 109 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.162 |
-0.399 |
| 110 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.162 |
0.395 |
| 111 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.161 |
-0.040 |