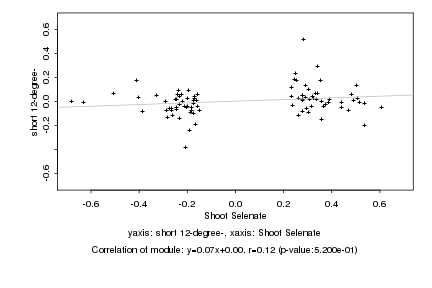
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Shoot Selenate |
Log2 signal ratio
short 12-degree- |
| 1 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.603 |
-0.046 |
| 2 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.533 |
-0.012 |
| 3 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.533 |
-0.196 |
| 4 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.512 |
-0.002 |
| 5 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.506 |
0.026 |
| 6 |
247800_at |
AT5G58570
|
unknown |
0.500 |
0.134 |
| 7 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.487 |
0.009 |
| 8 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.482 |
0.059 |
| 9 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.469 |
-0.070 |
| 10 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.439 |
-0.008 |
| 11 |
263515_at |
AT2G21640
|
unknown |
0.438 |
-0.048 |
| 12 |
259968_at |
AT1G76530
|
[Auxin efflux carrier family protein] |
0.389 |
0.022 |
| 13 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.385 |
-0.006 |
| 14 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
0.371 |
-0.023 |
| 15 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.363 |
-0.037 |
| 16 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.356 |
-0.149 |
| 17 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.354 |
0.004 |
| 18 |
260522_x_at |
AT2G41730
|
unknown |
0.350 |
0.182 |
| 19 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.340 |
0.075 |
| 20 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.338 |
0.299 |
| 21 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.333 |
0.025 |
| 22 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.332 |
0.073 |
| 23 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.322 |
0.032 |
| 24 |
248260_at |
AT5G53240
|
unknown |
0.317 |
0.049 |
| 25 |
249384_at |
AT5G39890
|
unknown |
0.313 |
-0.036 |
| 26 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.304 |
0.024 |
| 27 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.302 |
-0.086 |
| 28 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.302 |
0.104 |
| 29 |
252117_at |
AT3G51430
|
SSL5, STRICTOSIDINE SYNTHASE-LIKE 5, YLS2, YELLOW-LEAF-SPECIFIC GENE 2 |
0.295 |
-0.052 |
| 30 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.290 |
0.138 |
| 31 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.287 |
0.036 |
| 32 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.281 |
0.525 |
| 33 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.278 |
0.020 |
| 34 |
257421_at |
AT1G12030
|
unknown |
0.278 |
0.054 |
| 35 |
244945_at |
ATMG00110
|
ABCI2, ATP-binding cassette I2, CCB206, cytochrome C biogenesis 206 |
0.277 |
-0.078 |
| 36 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.275 |
0.016 |
| 37 |
244906_at |
ATMG00690
|
ORF240A |
0.275 |
0.057 |
| 38 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.261 |
-0.114 |
| 39 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.260 |
0.027 |
| 40 |
255259_at |
AT4G05020
|
NDB2, NAD(P)H dehydrogenase B2 |
0.251 |
0.178 |
| 41 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.246 |
0.237 |
| 42 |
264907_at |
AT2G17280
|
[Phosphoglycerate mutase family protein] |
0.243 |
0.192 |
| 43 |
249377_at |
AT5G40690
|
unknown |
0.234 |
-0.033 |
| 44 |
258452_at |
AT3G22370
|
AOX1A, alternative oxidase 1A, ATAOX1A, AtHSR3, HSR3, hyper-sensitivity-related 3 |
0.229 |
0.121 |
| 45 |
258982_at |
AT3G08870
|
LecRK-VI.1, L-type lectin receptor kinase VI.1 |
0.229 |
0.045 |
| 46 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
-0.684 |
0.006 |
| 47 |
266222_at |
AT2G28780
|
unknown |
-0.635 |
-0.005 |
| 48 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
-0.508 |
0.072 |
| 49 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
-0.414 |
0.181 |
| 50 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.403 |
0.037 |
| 51 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.388 |
-0.077 |
| 52 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.331 |
0.055 |
| 53 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.291 |
0.001 |
| 54 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.287 |
-0.073 |
| 55 |
259520_at |
AT1G12320
|
unknown |
-0.286 |
-0.127 |
| 56 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.277 |
-0.056 |
| 57 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.268 |
-0.072 |
| 58 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.267 |
-0.055 |
| 59 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.265 |
-0.115 |
| 60 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
-0.250 |
0.021 |
| 61 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.248 |
-0.060 |
| 62 |
260851_at |
AT1G21890
|
UMAMIT19, Usually multiple acids move in and out Transporters 19 |
-0.247 |
-0.047 |
| 63 |
246415_at |
AT5G17160
|
unknown |
-0.245 |
0.025 |
| 64 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.242 |
0.062 |
| 65 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.237 |
0.092 |
| 66 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.236 |
0.050 |
| 67 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.236 |
-0.018 |
| 68 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.235 |
-0.139 |
| 69 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
-0.227 |
0.060 |
| 70 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.221 |
0.006 |
| 71 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.212 |
-0.039 |
| 72 |
256096_at |
AT1G13650
|
unknown |
-0.208 |
-0.379 |
| 73 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
-0.205 |
-0.047 |
| 74 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.200 |
0.033 |
| 75 |
256222_at |
AT1G56210
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.200 |
-0.040 |
| 76 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
-0.197 |
0.099 |
| 77 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.193 |
-0.237 |
| 78 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.190 |
-0.091 |
| 79 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.184 |
-0.074 |
| 80 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.183 |
-0.047 |
| 81 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.177 |
0.010 |
| 82 |
267402_at |
AT2G26180
|
IQD6, IQ-domain 6 |
-0.176 |
-0.013 |
| 83 |
249060_at |
AT5G44560
|
VPS2.2 |
-0.176 |
-0.094 |
| 84 |
251021_at |
AT5G02140
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.174 |
0.027 |
| 85 |
259136_at |
AT3G05470
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.174 |
0.050 |
| 86 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.167 |
-0.187 |
| 87 |
248085_at |
AT5G55480
|
GDPDL4, Glycerophosphodiester phosphodiesterase (GDPD) like 4, GPDL1, glycerophosphodiesterase-like 1, SVL1, SHV3-like 1 |
-0.162 |
0.009 |
| 88 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.159 |
0.059 |
| 89 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.158 |
-0.034 |
| 90 |
259290_at |
AT3G11520
|
CYC2, CYCLIN 2, CYCB1;3, CYCLIN B1;3 |
-0.153 |
-0.069 |