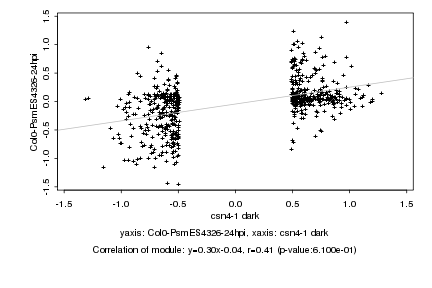
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
csn4-1 dark |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
1.275 |
0.148 |
| 2 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
1.197 |
0.002 |
| 3 |
265234_at |
AT2G07721
|
unknown |
1.197 |
0.051 |
| 4 |
248125_at |
AT5G54740
|
SESA5, seed storage albumin 5 |
1.181 |
-0.001 |
| 5 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
1.161 |
0.293 |
| 6 |
257336_at |
ATMG01410
|
ORF204, open reading frame 204 |
1.117 |
0.109 |
| 7 |
257330_at |
ATMG01290
|
ORF111C |
1.114 |
0.122 |
| 8 |
252661_at |
AT3G44450
|
unknown |
1.108 |
-0.080 |
| 9 |
267398_at |
AT2G44560
|
AtGH9B11, glycosyl hydrolase 9B11, GH9B11, glycosyl hydrolase 9B11 |
1.105 |
0.077 |
| 10 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
1.088 |
0.070 |
| 11 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
1.076 |
0.213 |
| 12 |
244947_at |
ATMG00130
|
ORF121A |
1.051 |
0.132 |
| 13 |
257323_at |
ATMG01200
|
ORF294 |
1.048 |
0.246 |
| 14 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
1.039 |
-0.082 |
| 15 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
1.014 |
0.627 |
| 16 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
1.005 |
-0.140 |
| 17 |
244911_at |
ATMG00820
|
ORF170 |
1.000 |
-0.002 |
| 18 |
244909_at |
ATMG00740
|
ORF100A |
0.998 |
0.044 |
| 19 |
264473_at |
AT1G67180
|
[zinc finger (C3HC4-type RING finger) family protein / BRCT domain-containing protein] |
0.972 |
-0.068 |
| 20 |
258327_at |
AT3G22640
|
PAP85 |
0.969 |
0.057 |
| 21 |
249532_at |
AT5G38780
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.965 |
0.781 |
| 22 |
257322_at |
ATMG01180
|
ORF111B |
0.965 |
0.037 |
| 23 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.965 |
0.252 |
| 24 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.965 |
1.398 |
| 25 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.955 |
0.027 |
| 26 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.944 |
0.464 |
| 27 |
257332_at |
ATMG01350
|
ORF145C |
0.937 |
0.083 |
| 28 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.925 |
-0.096 |
| 29 |
263769_at |
AT2G06390
|
transposable element gene |
0.924 |
-0.029 |
| 30 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.919 |
-0.209 |
| 31 |
257327_at |
ATMG01240
|
ORF100C |
0.908 |
0.141 |
| 32 |
257335_at |
ATMG01400
|
ORF105B |
0.907 |
0.027 |
| 33 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.906 |
0.181 |
| 34 |
244957_at |
ATMG00400
|
ORF157 |
0.904 |
0.090 |
| 35 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
0.890 |
0.317 |
| 36 |
244913_at |
ATMG00840
|
ORF121B |
0.886 |
-0.002 |
| 37 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.885 |
0.151 |
| 38 |
257321_at |
ATMG01130
|
ORF106F |
0.883 |
0.207 |
| 39 |
244931_at |
ATMG00630
|
ORF110B |
0.881 |
0.040 |
| 40 |
246025_at |
AT5G21150
|
AGO9, ARGONAUTE 9 |
0.877 |
0.156 |
| 41 |
252010_at |
AT3G52740
|
unknown |
0.875 |
0.076 |
| 42 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.866 |
0.702 |
| 43 |
260459_at |
AT1G68240
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.864 |
-0.028 |
| 44 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
0.863 |
-0.307 |
| 45 |
263819_x_at |
AT2G10140
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 1.7e-140 P-value blast match to GB:CAA40555 TNP2 (CACTA-element) (Antirrhinum majus)] |
0.860 |
0.086 |
| 46 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.859 |
-0.105 |
| 47 |
244941_at |
ATMG00010
|
ORF153A |
0.857 |
0.032 |
| 48 |
266046_at |
AT2G07728
|
unknown |
0.857 |
-0.036 |
| 49 |
263881_at |
AT2G21820
|
unknown |
0.850 |
0.361 |
| 50 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.847 |
-0.253 |
| 51 |
265242_at |
AT2G07705
|
unknown |
0.846 |
0.092 |
| 52 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
0.845 |
0.083 |
| 53 |
266393_at |
AT2G41260
|
ATM17, M17 |
0.844 |
0.014 |
| 54 |
244994_at |
ATCG01010
|
NDHF |
0.842 |
0.000 |
| 55 |
244942_at |
ATMG00050
|
ORF131 |
0.842 |
0.096 |
| 56 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
0.835 |
0.088 |
| 57 |
250882_at |
AT5G04000
|
unknown |
0.832 |
0.111 |
| 58 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.827 |
0.042 |
| 59 |
257574_at |
AT3G20710
|
[F-box family protein] |
0.823 |
0.021 |
| 60 |
244945_at |
ATMG00110
|
ABCI2, ATP-binding cassette I2, CCB206, cytochrome C biogenesis 206 |
0.817 |
0.180 |
| 61 |
257456_at |
AT2G18120
|
SRS4, SHI-related sequence 4 |
0.814 |
-0.028 |
| 62 |
256538_x_at |
AT3G32900
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT3G47260.1)] |
0.808 |
0.060 |
| 63 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.806 |
0.132 |
| 64 |
255376_x_at |
AT4G03790
|
[gypsy-like retrotransposon family (Athila), has a 2.2e-286 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.798 |
0.076 |
| 65 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
0.797 |
0.033 |
| 66 |
257777_x_at |
AT3G29210
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
0.797 |
0.018 |
| 67 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.796 |
0.274 |
| 68 |
266849_at |
AT2G25940
|
ALPHA-VPE, alpha-vacuolar processing enzyme, ALPHAVPE |
0.795 |
0.159 |
| 69 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.795 |
0.038 |
| 70 |
249454_at |
AT5G39520
|
unknown |
0.783 |
0.805 |
| 71 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
0.780 |
0.128 |
| 72 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.775 |
-0.162 |
| 73 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.774 |
-0.035 |
| 74 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
0.769 |
0.050 |
| 75 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.769 |
0.262 |
| 76 |
264494_at |
AT1G27461
|
unknown |
0.768 |
0.110 |
| 77 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.764 |
0.636 |
| 78 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
0.762 |
0.051 |
| 79 |
244901_at |
ATMG00640
|
ORF25 |
0.760 |
0.361 |
| 80 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
0.759 |
0.777 |
| 81 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.757 |
0.405 |
| 82 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.755 |
0.378 |
| 83 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
0.754 |
0.169 |
| 84 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.753 |
0.167 |
| 85 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.752 |
1.142 |
| 86 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.751 |
-0.527 |
| 87 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
0.743 |
-0.053 |
| 88 |
265237_s_at |
ATMG00470
|
ORF122A |
0.743 |
0.061 |
| 89 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
0.741 |
0.109 |
| 90 |
257110_x_at |
AT3G30440
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
0.740 |
0.131 |
| 91 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.740 |
-0.504 |
| 92 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.739 |
0.226 |
| 93 |
259615_at |
AT1G47980
|
unknown |
0.739 |
0.037 |
| 94 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
0.738 |
0.004 |
| 95 |
244948_at |
ATMG00140
|
ORF167 |
0.735 |
-0.007 |
| 96 |
244907_at |
ATMG00710
|
ORF120 |
0.733 |
0.036 |
| 97 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.730 |
0.946 |
| 98 |
259489_at |
AT1G15790
|
unknown |
0.729 |
0.574 |
| 99 |
244921_s_at |
ATMG01000
|
ORF114 |
0.727 |
0.101 |
| 100 |
255120_x_at |
AT4G08890
|
[Mutator-like transposase family, has a 5.6e-82 P-value blast match to O65231 /281-442 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
0.727 |
0.038 |
| 101 |
244923_s_at |
ATMG01020
|
ORF153B |
0.726 |
0.035 |
| 102 |
244903_at |
ATMG00660
|
ORF149 |
0.719 |
0.177 |
| 103 |
251535_at |
AT3G58540
|
unknown |
0.718 |
0.092 |
| 104 |
261619_x_at |
AT1G33130
|
[CACTA-like transposase family (Ptta/En/Spm), has a 1.3e-125 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.715 |
0.038 |
| 105 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.714 |
-0.182 |
| 106 |
245126_at |
AT2G47460
|
ATMYB12, MYB DOMAIN PROTEIN 12, MYB12, myb domain protein 12, PFG1, PRODUCTION OF FLAVONOL GLYCOSIDES 1 |
0.710 |
0.128 |
| 107 |
244999_at |
ATCG00190
|
RPOB, RNA polymerase subunit beta |
0.709 |
-0.262 |
| 108 |
266015_at |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
0.704 |
-0.166 |
| 109 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.704 |
0.552 |
| 110 |
261235_x_at |
AT1G32840
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
0.701 |
0.045 |
| 111 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.701 |
0.864 |
| 112 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
0.700 |
-0.603 |
| 113 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.698 |
0.505 |
| 114 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
0.696 |
0.567 |
| 115 |
253781_at |
AT4G28580
|
ATMGT5, ARABIDOPSIS THALIANA MAGNESIUM TRANSPORT 5, MGT5, magnesium transport 5, MRS2-6 |
0.687 |
0.065 |
| 116 |
263515_at |
AT2G21640
|
unknown |
0.685 |
0.587 |
| 117 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
0.684 |
0.072 |
| 118 |
267036_at |
AT2G38465
|
unknown |
0.684 |
0.202 |
| 119 |
254565_at |
AT4G19130
|
[Replication factor-A protein 1-related] |
0.679 |
-0.064 |
| 120 |
255351_x_at |
AT4G03850
|
[gypsy-like retrotransposon family (Athila), has a 1.8e-131 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.676 |
0.038 |
| 121 |
244955_at |
ATMG00320
|
ORF127 |
0.675 |
0.092 |
| 122 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.669 |
0.466 |
| 123 |
258307_x_at |
AT3G30560
|
[similar to AT hook motif-containing protein-related [Arabidopsis thaliana] (TAIR:AT1G35940.1)] |
0.665 |
0.023 |
| 124 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.664 |
0.156 |
| 125 |
244958_at |
ATMG00450
|
ORF106B |
0.661 |
0.017 |
| 126 |
260517_at |
AT1G51420
|
ATSPP1, SUCROSE-PHOSPHATASE 1, SPP1, sucrose-phosphatase 1 |
0.657 |
0.050 |
| 127 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
0.654 |
-0.035 |
| 128 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.652 |
0.433 |
| 129 |
244917_at |
ATMG00870
|
ORF184 |
0.652 |
0.103 |
| 130 |
253494_at |
AT4G31830
|
unknown |
0.651 |
0.058 |
| 131 |
251354_at |
AT3G61090
|
[Putative endonuclease or glycosyl hydrolase] |
0.648 |
0.069 |
| 132 |
255197_x_at |
AT4G07450
|
unknown |
0.646 |
0.007 |
| 133 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
0.640 |
-0.066 |
| 134 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.638 |
0.027 |
| 135 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.637 |
0.355 |
| 136 |
259386_at |
AT1G13400
|
JGL, JAGGED-LIKE, NUB, NUBBIN |
0.636 |
0.043 |
| 137 |
257334_at |
ATMG01370
|
ORF111D |
0.633 |
-0.052 |
| 138 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
0.631 |
0.086 |
| 139 |
264752_at |
AT1G23010
|
LPR1, Low Phosphate Root1 |
0.625 |
0.215 |
| 140 |
251485_at |
AT3G59550
|
ATRAD21.2, SISTER CHROMATID COHESION 1 PROTEIN 3, ATSYN3, SYN3 |
0.625 |
0.001 |
| 141 |
263214_at |
AT1G30660
|
[nucleic acid binding] |
0.623 |
0.031 |
| 142 |
257111_x_at |
AT3G30450
|
[similar to heat shock protein binding [Arabidopsis thaliana] (TAIR:AT3G30450.1)] |
0.620 |
0.019 |
| 143 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
0.619 |
0.732 |
| 144 |
246582_at |
AT1G31750
|
[proline-rich family protein] |
0.616 |
0.427 |
| 145 |
253866_at |
AT4G27480
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.613 |
0.197 |
| 146 |
246640_x_at |
AT5G34900
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT3G47260.1)] |
0.612 |
-0.042 |
| 147 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.612 |
-0.138 |
| 148 |
253942_at |
AT4G27010
|
EMB2788, EMBRYO DEFECTIVE 2788 |
0.611 |
-0.216 |
| 149 |
254287_at |
AT4G22960
|
unknown |
0.609 |
0.085 |
| 150 |
248752_at |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
0.609 |
0.063 |
| 151 |
248494_at |
AT5G50770
|
AtHSD6, hydroxysteroid dehydrogenase 6, HSD6, hydroxysteroid dehydrogenase 6 |
0.608 |
0.052 |
| 152 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.604 |
0.027 |
| 153 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
0.603 |
0.028 |
| 154 |
265296_at |
AT2G14060
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.602 |
-0.057 |
| 155 |
262106_at |
AT1G02970
|
ATWEE1, WEE1, WEE1 kinase homolog |
0.602 |
-0.011 |
| 156 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.601 |
0.121 |
| 157 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
0.598 |
0.214 |
| 158 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.598 |
0.797 |
| 159 |
247857_at |
AT5G58400
|
[Peroxidase superfamily protein] |
0.598 |
0.124 |
| 160 |
261588_at |
AT1G01670
|
[RING/U-box superfamily protein] |
0.597 |
0.159 |
| 161 |
255840_at |
AT2G33520
|
unknown |
0.596 |
0.131 |
| 162 |
251499_at |
AT3G59100
|
ATGSL11, glucan synthase-like 11, gsl11, GSL11, glucan synthase-like 11 |
0.596 |
0.220 |
| 163 |
251730_at |
AT3G56330
|
[N2,N2-dimethylguanosine tRNA methyltransferase] |
0.593 |
-0.293 |
| 164 |
253796_at |
AT4G28460
|
unknown |
0.591 |
0.849 |
| 165 |
261511_at |
AT1G71770
|
PAB5, poly(A)-binding protein 5 |
0.588 |
0.060 |
| 166 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.587 |
1.028 |
| 167 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.587 |
0.734 |
| 168 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.585 |
0.749 |
| 169 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
0.583 |
0.466 |
| 170 |
256460_at |
AT1G36240
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.580 |
0.048 |
| 171 |
266040_at |
AT2G07738
|
unknown |
0.580 |
0.014 |
| 172 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
0.579 |
0.368 |
| 173 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
0.579 |
-0.205 |
| 174 |
251202_at |
AT3G63040
|
unknown |
0.579 |
0.137 |
| 175 |
246054_at |
AT5G08360
|
unknown |
0.578 |
0.058 |
| 176 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
0.578 |
-0.284 |
| 177 |
253240_at |
AT4G34510
|
KCS17, 3-ketoacyl-CoA synthase 17 |
0.577 |
0.101 |
| 178 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
0.576 |
0.197 |
| 179 |
245000_at |
ATCG00210
|
YCF6 |
0.575 |
-0.022 |
| 180 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.575 |
0.671 |
| 181 |
249129_at |
AT5G43080
|
CYCA3;1, Cyclin A3;1 |
0.575 |
-0.209 |
| 182 |
255094_at |
AT4G08590
|
ORL1, ORTH-LIKE 1, ORL1, ORTHRUS-LIKE 1, ORTHL, ORTHRUS-like, VIM6, VARIANT IN METHYLATION 6 |
0.574 |
-0.157 |
| 183 |
256188_at |
AT1G30160
|
unknown |
0.573 |
0.031 |
| 184 |
267118_at |
AT2G32590
|
EMB2795, EMBRYO DEFECTIVE 2795 |
0.572 |
-0.036 |
| 185 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
0.570 |
-0.059 |
| 186 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.570 |
0.194 |
| 187 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.569 |
-0.071 |
| 188 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
0.569 |
0.196 |
| 189 |
247290_at |
AT5G64450
|
unknown |
0.568 |
0.177 |
| 190 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.566 |
0.412 |
| 191 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.564 |
0.694 |
| 192 |
254479_at |
AT4G20350
|
[oxidoreductases] |
0.562 |
-0.024 |
| 193 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.560 |
0.134 |
| 194 |
244906_at |
ATMG00690
|
ORF240A |
0.557 |
0.037 |
| 195 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.556 |
0.569 |
| 196 |
245522_at |
AT4G15890
|
[binding] |
0.555 |
0.137 |
| 197 |
256425_at |
AT1G33560
|
ADR1, ACTIVATED DISEASE RESISTANCE 1 |
0.555 |
0.379 |
| 198 |
247167_at |
AT5G65850
|
[F-box and associated interaction domains-containing protein] |
0.554 |
0.015 |
| 199 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
0.553 |
0.308 |
| 200 |
251273_at |
AT3G61960
|
[Protein kinase superfamily protein] |
0.553 |
0.313 |
| 201 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
0.552 |
0.098 |
| 202 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
0.551 |
0.459 |
| 203 |
245560_at |
AT4G15480
|
UGT84A1 |
0.550 |
0.112 |
| 204 |
267081_at |
AT2G41210
|
PIP5K5, phosphatidylinositol- 4-phosphate 5-kinase 5 |
0.549 |
0.075 |
| 205 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
0.548 |
0.525 |
| 206 |
258652_at |
AT3G09910
|
ATRAB18C, ATRABC2B, RAB GTPase homolog C2B, RABC2b, RAB GTPase homolog C2B |
0.544 |
0.966 |
| 207 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
0.544 |
0.237 |
| 208 |
248225_at |
AT5G53740
|
unknown |
0.543 |
-0.012 |
| 209 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
0.543 |
-0.045 |
| 210 |
257317_at |
ATMG01060
|
ORF107G |
0.542 |
0.006 |
| 211 |
251852_at |
AT3G54750
|
unknown |
0.542 |
-0.161 |
| 212 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.541 |
-0.468 |
| 213 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.541 |
1.068 |
| 214 |
253689_at |
AT4G29770
|
unknown |
0.540 |
0.082 |
| 215 |
250620_at |
AT5G07190
|
ATS3, seed gene 3 |
0.540 |
0.101 |
| 216 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.540 |
0.015 |
| 217 |
254729_at |
AT4G13700
|
ATPAP23, PAP23, purple acid phosphatase 23 |
0.539 |
0.015 |
| 218 |
248484_at |
AT5G51030
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.539 |
0.301 |
| 219 |
257306_at |
AT3G30200
|
[Plant transposase (Ptta/En/Spm family)] |
0.537 |
0.089 |
| 220 |
249747_at |
AT5G24600
|
unknown |
0.536 |
0.191 |
| 221 |
255594_at |
AT4G01660
|
ABC1, ABC transporter 1, ATABC1, ABC transporter 1, ATATH10 |
0.535 |
0.083 |
| 222 |
265847_at |
AT2G35750
|
unknown |
0.534 |
-0.027 |
| 223 |
260544_at |
AT2G43540
|
unknown |
0.534 |
0.347 |
| 224 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
0.533 |
0.009 |
| 225 |
262321_at |
AT1G27570
|
[phosphatidylinositol 3- and 4-kinase family protein] |
0.531 |
0.068 |
| 226 |
258117_at |
AT3G14700
|
[SART-1 family] |
0.530 |
0.550 |
| 227 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.529 |
0.643 |
| 228 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.528 |
0.174 |
| 229 |
257813_at |
AT3G25100
|
CDC45, cell division cycle 45 |
0.528 |
0.106 |
| 230 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
0.527 |
0.120 |
| 231 |
247196_at |
AT5G65510
|
AIL7, AINTEGUMENTA-like 7, PLT7, PLETHORA 7 |
0.527 |
0.075 |
| 232 |
257086_at |
AT3G20490
|
unknown protein; Has 754 Blast hits to 165 proteins in 64 species: Archae - 0; Bacteria - 48; Metazoa - 26; Fungi - 25; Plants - 36; Viruses - 0; Other Eukaryotes - 619 (source: NCBI BLink). |
0.526 |
0.156 |
| 233 |
257850_at |
AT3G13065
|
SRF4, STRUBBELIG-receptor family 4 |
0.525 |
-0.059 |
| 234 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.524 |
0.312 |
| 235 |
252283_at |
AT3G48960
|
[Ribosomal protein L13e family protein] |
0.524 |
-0.078 |
| 236 |
244908_at |
ATMG00720
|
ORF107D |
0.522 |
0.024 |
| 237 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
0.521 |
-0.015 |
| 238 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
0.520 |
-0.084 |
| 239 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.519 |
0.139 |
| 240 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.519 |
0.750 |
| 241 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.519 |
-0.119 |
| 242 |
267623_at |
AT2G39650
|
unknown |
0.519 |
0.455 |
| 243 |
256748_x_at |
AT3G30396
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.1e-39 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.518 |
0.054 |
| 244 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.518 |
0.373 |
| 245 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.517 |
0.666 |
| 246 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.517 |
0.034 |
| 247 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.516 |
-0.219 |
| 248 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
0.516 |
-0.251 |
| 249 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.515 |
-0.061 |
| 250 |
244929_at |
ATMG00580
|
NAD4, NADH dehydrogenase subunit 4 |
0.514 |
0.090 |
| 251 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.514 |
0.131 |
| 252 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
0.514 |
1.008 |
| 253 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.512 |
0.721 |
| 254 |
257346_at |
AT3G30846
|
[gypsy-like retrotransposon family, has a 1.0e-303 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
0.512 |
0.063 |
| 255 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.511 |
-0.189 |
| 256 |
252962_at |
AT4G38780
|
[Pre-mRNA-processing-splicing factor] |
0.510 |
0.024 |
| 257 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.509 |
0.770 |
| 258 |
253506_at |
AT4G31980
|
unknown |
0.509 |
0.074 |
| 259 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.506 |
0.620 |
| 260 |
246973_at |
AT5G24970
|
[Protein kinase superfamily protein] |
0.506 |
0.028 |
| 261 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.506 |
0.132 |
| 262 |
244995_at |
ATCG00150
|
ATPI |
0.506 |
-0.373 |
| 263 |
245197_at |
AT1G67800
|
[Copine (Calcium-dependent phospholipid-binding protein) family] |
0.505 |
0.348 |
| 264 |
252208_at |
AT3G50380
|
[INVOLVED IN: protein localization] |
0.505 |
0.162 |
| 265 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.504 |
0.304 |
| 266 |
265892_at |
AT2G15020
|
unknown |
0.504 |
-0.712 |
| 267 |
266151_x_at |
AT2G12300
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.8e-41 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.503 |
0.032 |
| 268 |
246132_at |
AT5G20850
|
ATRAD51, RAD51 |
0.503 |
-0.014 |
| 269 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
0.503 |
0.729 |
| 270 |
258982_at |
AT3G08870
|
LecRK-VI.1, L-type lectin receptor kinase VI.1 |
0.503 |
0.303 |
| 271 |
265897_at |
AT2G25680
|
MOT1, molybdate transporter 1 |
0.502 |
-0.036 |
| 272 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.501 |
1.019 |
| 273 |
247788_at |
AT5G58730
|
Mik, myo-inositol kinase |
0.500 |
0.343 |
| 274 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.500 |
1.236 |
| 275 |
249321_at |
AT5G40920
|
[pseudogene] |
0.499 |
0.078 |
| 276 |
257994_at |
AT3G19920
|
unknown |
0.499 |
-0.024 |
| 277 |
247672_at |
AT5G60220
|
TET4, tetraspanin4 |
0.499 |
0.067 |
| 278 |
250722_at |
AT5G06190
|
unknown |
0.499 |
0.445 |
| 279 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
0.499 |
0.227 |
| 280 |
263853_at |
AT2G04440
|
[MutT/nudix family protein] |
0.498 |
0.124 |
| 281 |
263122_at |
AT1G78510
|
AtSPS1, SPS1, solanesyl diphosphate synthase 1 |
0.498 |
0.012 |
| 282 |
258242_at |
AT3G27640
|
[Transducin/WD40 repeat-like superfamily protein] |
0.497 |
0.073 |
| 283 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.496 |
0.025 |
| 284 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.496 |
-0.674 |
| 285 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.495 |
0.597 |
| 286 |
256230_at |
AT3G12340
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.495 |
0.003 |
| 287 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.495 |
0.744 |
| 288 |
265241_at |
AT2G07693
|
[copia-like retrotransposon family, has a 6.2e-16 P-value blast match to GB:CAA37924 orf 2 (Ty1_Copia-element) (Arabidopsis thaliana)] |
0.495 |
0.169 |
| 289 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.494 |
0.241 |
| 290 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.494 |
0.343 |
| 291 |
250372_at |
AT5G11460
|
unknown |
0.493 |
-0.165 |
| 292 |
263520_at |
AT2G42640
|
[Mitogen activated protein kinase kinase kinase-related] |
0.492 |
0.036 |
| 293 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.492 |
0.603 |
| 294 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
0.490 |
0.392 |
| 295 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.489 |
0.701 |
| 296 |
252383_at |
AT3G47780
|
ABCA7, ATP-binding cassette A7, ATATH6, A. THALIANA ABC2 HOMOLOG 6, ATH6, ABC2 homolog 6 |
0.488 |
0.718 |
| 297 |
254917_at |
AT4G11350
|
unknown |
0.488 |
0.367 |
| 298 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.487 |
0.901 |
| 299 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.485 |
-0.831 |
| 300 |
266061_at |
AT2G18720
|
[Translation elongation factor EF1A/initiation factor IF2gamma family protein] |
0.485 |
0.125 |
| 301 |
267459_at |
AT2G33850
|
unknown |
-1.314 |
0.044 |
| 302 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-1.291 |
0.065 |
| 303 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-1.164 |
-1.150 |
| 304 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-1.096 |
-0.458 |
| 305 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.076 |
-0.645 |
| 306 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.038 |
-0.080 |
| 307 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
-1.029 |
-0.577 |
| 308 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-1.017 |
-0.639 |
| 309 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-1.012 |
-0.735 |
| 310 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
-1.007 |
0.038 |
| 311 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-1.004 |
-0.721 |
| 312 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.981 |
-0.135 |
| 313 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.976 |
-1.036 |
| 314 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.966 |
-0.096 |
| 315 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.962 |
-0.308 |
| 316 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.961 |
-0.193 |
| 317 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.955 |
-0.027 |
| 318 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.943 |
-1.026 |
| 319 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.941 |
-0.250 |
| 320 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.939 |
0.129 |
| 321 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.936 |
-0.013 |
| 322 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.934 |
0.159 |
| 323 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.932 |
-0.100 |
| 324 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.931 |
-0.802 |
| 325 |
267545_at |
AT2G32690
|
ATGRP23, GLYCINE-RICH PROTEIN 23, GRP23, glycine-rich protein 23 |
-0.923 |
-0.400 |
| 326 |
247474_at |
AT5G62280
|
unknown |
-0.912 |
-0.600 |
| 327 |
262693_at |
AT1G62780
|
unknown |
-0.901 |
-0.471 |
| 328 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.898 |
-0.225 |
| 329 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.893 |
-1.046 |
| 330 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.888 |
0.080 |
| 331 |
247878_at |
AT5G57760
|
unknown |
-0.876 |
-0.217 |
| 332 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.874 |
-1.106 |
| 333 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.865 |
-1.009 |
| 334 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.859 |
0.499 |
| 335 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
-0.859 |
0.068 |
| 336 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.849 |
-0.108 |
| 337 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.839 |
-0.994 |
| 338 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.835 |
0.441 |
| 339 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.835 |
-0.432 |
| 340 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
-0.833 |
0.005 |
| 341 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.825 |
-0.466 |
| 342 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.825 |
-0.716 |
| 343 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.821 |
-0.786 |
| 344 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.808 |
-0.555 |
| 345 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.802 |
-0.240 |
| 346 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.799 |
0.130 |
| 347 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.797 |
-0.768 |
| 348 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.795 |
-0.503 |
| 349 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.789 |
0.061 |
| 350 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.786 |
-0.391 |
| 351 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.783 |
-0.574 |
| 352 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.776 |
-0.447 |
| 353 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.769 |
0.118 |
| 354 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.768 |
-0.228 |
| 355 |
246583_at |
AT5G14720
|
[Protein kinase superfamily protein] |
-0.767 |
0.052 |
| 356 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.766 |
0.960 |
| 357 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.765 |
0.126 |
| 358 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.764 |
-0.989 |
| 359 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
-0.762 |
-0.499 |
| 360 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.758 |
-0.310 |
| 361 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.754 |
-0.036 |
| 362 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
-0.751 |
-0.037 |
| 363 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.751 |
-0.797 |
| 364 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
-0.750 |
-0.793 |
| 365 |
246860_at |
AT5G25840
|
unknown |
-0.748 |
-0.141 |
| 366 |
264910_at |
AT1G61100
|
[disease resistance protein (TIR class), putative] |
-0.746 |
0.098 |
| 367 |
262796_at |
AT1G20850
|
XCP2, xylem cysteine peptidase 2 |
-0.742 |
0.012 |
| 368 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
-0.742 |
-0.911 |
| 369 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.741 |
-0.768 |
| 370 |
255028_at |
AT4G09890
|
unknown |
-0.730 |
-0.421 |
| 371 |
259373_at |
AT1G69160
|
unknown |
-0.729 |
-0.617 |
| 372 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.727 |
-0.186 |
| 373 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.725 |
-0.886 |
| 374 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.723 |
-0.136 |
| 375 |
260012_at |
AT1G67865
|
unknown |
-0.716 |
0.139 |
| 376 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.715 |
0.407 |
| 377 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.713 |
-0.839 |
| 378 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.713 |
0.274 |
| 379 |
267279_at |
AT2G19460
|
unknown |
-0.711 |
-0.033 |
| 380 |
250327_at |
AT5G12050
|
unknown |
-0.709 |
-1.145 |
| 381 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.709 |
0.041 |
| 382 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.707 |
-0.991 |
| 383 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
-0.702 |
0.100 |
| 384 |
248382_at |
AT5G51890
|
[Peroxidase superfamily protein] |
-0.702 |
0.054 |
| 385 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.699 |
0.237 |
| 386 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
-0.699 |
0.089 |
| 387 |
248801_at |
AT5G47370
|
HAT2 |
-0.695 |
-0.107 |
| 388 |
250300_at |
AT5G11890
|
EMB3135, EMBRYO DEFECTIVE 3135 |
-0.694 |
-0.299 |
| 389 |
253062_at |
AT4G37590
|
MEL1, MAB4/ENP/NPY1-LIKE 1, NPY5, NAKED PINS IN YUC MUTANTS 5 |
-0.692 |
-0.422 |
| 390 |
256021_at |
AT1G58270
|
ZW9 |
-0.691 |
-0.107 |
| 391 |
260445_at |
AT1G68370
|
ARG1, ALTERED RESPONSE TO GRAVITY 1 |
-0.691 |
0.267 |
| 392 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
-0.689 |
0.707 |
| 393 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.688 |
0.132 |
| 394 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
-0.685 |
0.532 |
| 395 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
-0.684 |
0.117 |
| 396 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.683 |
-0.103 |
| 397 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.682 |
0.102 |
| 398 |
267481_at |
AT2G02780
|
Leucine-rich repeat protein kinase family protein |
-0.680 |
-0.456 |
| 399 |
264343_at |
AT1G11850
|
unknown |
-0.678 |
-0.573 |
| 400 |
261292_at |
AT1G36940
|
unknown |
-0.674 |
-0.058 |
| 401 |
250939_at |
AT5G03040
|
iqd2, IQ-domain 2 |
-0.673 |
-0.177 |
| 402 |
249109_at |
AT5G43700
|
ATAUX2-11, AUXIN INDUCIBLE 2-11, IAA4, indole-3-acetic acid inducible 4 |
-0.670 |
-0.435 |
| 403 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.668 |
-0.247 |
| 404 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.667 |
0.184 |
| 405 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.667 |
-0.949 |
| 406 |
261502_at |
AT1G14440
|
AtHB31, homeobox protein 31, FTM2, FLORAL TRANSITION AT THE MERISTEM2, HB31, homeobox protein 31, ZHD4, ZINC FINGER HOMEODOMAIN 4 |
-0.665 |
-0.213 |
| 407 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.658 |
-0.281 |
| 408 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
-0.657 |
0.067 |
| 409 |
250809_at |
AT5G05140
|
[Transcription elongation factor (TFIIS) family protein] |
-0.654 |
0.617 |
| 410 |
245076_at |
AT2G23170
|
GH3.3 |
-0.654 |
0.849 |
| 411 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.653 |
0.121 |
| 412 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.651 |
-0.406 |
| 413 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.650 |
-0.946 |
| 414 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.648 |
-0.935 |
| 415 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
-0.648 |
0.134 |
| 416 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.647 |
-0.449 |
| 417 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.645 |
-0.641 |
| 418 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.643 |
-0.180 |
| 419 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
-0.643 |
-0.200 |
| 420 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.643 |
-0.123 |
| 421 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.642 |
0.073 |
| 422 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.641 |
-0.890 |
| 423 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.640 |
-0.278 |
| 424 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.640 |
-0.454 |
| 425 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.639 |
-0.777 |
| 426 |
264900_at |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
-0.639 |
-0.516 |
| 427 |
264990_at |
AT1G27210
|
[ARM repeat superfamily protein] |
-0.637 |
-0.212 |
| 428 |
250464_at |
AT5G10040
|
unknown |
-0.636 |
0.154 |
| 429 |
252647_at |
AT3G44620
|
[protein tyrosine phosphatases] |
-0.634 |
0.123 |
| 430 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-0.633 |
0.084 |
| 431 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
-0.633 |
-0.133 |
| 432 |
258472_at |
AT3G06080
|
TBL10, TRICHOME BIREFRINGENCE-LIKE 10 |
-0.631 |
-0.119 |
| 433 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
-0.630 |
-0.238 |
| 434 |
262220_at |
AT1G74740
|
ATCPK30, CDPK1A, CALCIUM-DEPENDENT PROTEIN KINASE 1A, CPK30, calcium-dependent protein kinase 30 |
-0.627 |
0.303 |
| 435 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
-0.625 |
-0.001 |
| 436 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.624 |
0.077 |
| 437 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.622 |
-0.105 |
| 438 |
261611_at |
AT1G49730
|
[Protein kinase superfamily protein] |
-0.616 |
-0.474 |
| 439 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
-0.614 |
-0.488 |
| 440 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.614 |
-0.551 |
| 441 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.614 |
0.049 |
| 442 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.613 |
0.141 |
| 443 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
-0.613 |
0.074 |
| 444 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.613 |
-0.588 |
| 445 |
248772_at |
AT5G47800
|
[Phototropic-responsive NPH3 family protein] |
-0.611 |
-0.180 |
| 446 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.609 |
-0.392 |
| 447 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.609 |
0.147 |
| 448 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.609 |
-0.895 |
| 449 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.609 |
-1.045 |
| 450 |
266990_at |
AT2G39190
|
ATATH8 |
-0.608 |
-0.245 |
| 451 |
258468_at |
AT3G06070
|
unknown |
-0.608 |
-0.771 |
| 452 |
265011_at |
AT1G24490
|
ALB4, ALBINA 4, ARTEMIS, ARABIDOPSIS THALIANA ENVELOPE MEMBRANE INTEGRASE |
-0.607 |
0.004 |
| 453 |
247713_at |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.606 |
-0.053 |
| 454 |
262951_at |
AT1G75500
|
UMAMIT5, Usually multiple acids move in and out Transporters 5, WAT1, Walls Are Thin 1 |
-0.606 |
-0.437 |
| 455 |
245645_at |
AT1G24764
|
ATMAP70-2, microtubule-associated proteins 70-2, MAP70-2, microtubule-associated proteins 70-2 |
-0.604 |
-0.181 |
| 456 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.603 |
0.091 |
| 457 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.603 |
-1.425 |
| 458 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.602 |
0.080 |
| 459 |
260867_at |
AT1G43790
|
TED6, tracheary element differentiation-related 6 |
-0.601 |
-0.020 |
| 460 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.600 |
-0.492 |
| 461 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.600 |
0.034 |
| 462 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.600 |
-0.715 |
| 463 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.598 |
0.029 |
| 464 |
266117_at |
AT2G02170
|
[Remorin family protein] |
-0.597 |
-0.430 |
| 465 |
257971_at |
AT3G27530
|
GC6, golgin candidate 6, MAG4, MAIGO 4 |
-0.597 |
0.196 |
| 466 |
254169_at |
AT4G24290
|
[MAC/Perforin domain-containing protein] |
-0.595 |
0.226 |
| 467 |
263457_at |
AT2G22300
|
CAMTA3, CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 3, SR1, signal responsive 1 |
-0.594 |
0.370 |
| 468 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.592 |
-0.412 |
| 469 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
-0.590 |
0.554 |
| 470 |
257828_at |
AT3G26670
|
unknown |
-0.587 |
0.403 |
| 471 |
245792_at |
AT1G32100
|
ATPRR1, PRR1, pinoresinol reductase 1 |
-0.586 |
0.079 |
| 472 |
263114_at |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
-0.583 |
-0.256 |
| 473 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.581 |
-0.099 |
| 474 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
-0.579 |
-0.762 |
| 475 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.579 |
0.075 |
| 476 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.579 |
0.285 |
| 477 |
259877_at |
AT1G76710
|
ASHH1, ASH1 RELATED PROTEIN 1, ASHH1, ASH1-RELATED PROTEIN 1, SDG26, SET domain group 26 |
-0.579 |
0.018 |
| 478 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.579 |
-0.437 |
| 479 |
251050_at |
AT5G02440
|
unknown |
-0.578 |
0.010 |
| 480 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.578 |
0.020 |
| 481 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.574 |
-0.524 |
| 482 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.574 |
0.052 |
| 483 |
259166_at |
AT3G01670
|
AtSEOR2, Arabidopsis thaliana sieve element occlusion-related 2, SEOa, sieve element occlusion a, SEOR2, sieve element occlusion-related 2 |
-0.573 |
0.047 |
| 484 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
-0.572 |
-0.215 |
| 485 |
253606_at |
AT4G30530
|
GGP1, gamma-glutamyl peptidase 1 |
-0.571 |
0.129 |
| 486 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.569 |
0.051 |
| 487 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.565 |
-0.198 |
| 488 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.565 |
-0.802 |
| 489 |
252149_at |
AT3G51290
|
APSR1, Altered Phosphate Starvation Response 1 |
-0.565 |
0.091 |
| 490 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.565 |
-0.023 |
| 491 |
266324_at |
AT2G46710
|
ROPGAP3, ROP guanosine triphosphatase (GTPase)-activating protein 3 |
-0.565 |
-0.663 |
| 492 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.563 |
0.040 |
| 493 |
246975_at |
AT5G24890
|
unknown |
-0.563 |
0.127 |
| 494 |
247754_at |
AT5G59080
|
unknown |
-0.563 |
-0.593 |
| 495 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
-0.561 |
-0.330 |
| 496 |
261175_at |
AT1G04800
|
[glycine-rich protein] |
-0.560 |
-0.172 |
| 497 |
252215_at |
AT3G50240
|
KICP-02 |
-0.559 |
-0.625 |
| 498 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.559 |
-0.914 |
| 499 |
258196_at |
AT3G13980
|
unknown |
-0.558 |
-0.391 |
| 500 |
258935_at |
AT3G10120
|
unknown |
-0.558 |
0.026 |
| 501 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.557 |
-0.732 |
| 502 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.557 |
-0.307 |
| 503 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.557 |
-0.242 |
| 504 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.556 |
-0.566 |
| 505 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.555 |
-0.256 |
| 506 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.554 |
-0.628 |
| 507 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.552 |
-0.384 |
| 508 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.552 |
-0.305 |
| 509 |
251829_at |
AT3G55020
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
-0.551 |
0.016 |
| 510 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.550 |
-0.877 |
| 511 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.549 |
0.098 |
| 512 |
266125_at |
AT2G45050
|
GATA2, GATA transcription factor 2 |
-0.549 |
-0.066 |
| 513 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.549 |
-0.976 |
| 514 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.548 |
0.141 |
| 515 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.548 |
-0.093 |
| 516 |
255876_at |
AT2G40480
|
unknown |
-0.546 |
-0.562 |
| 517 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.546 |
-0.710 |
| 518 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
-0.545 |
-0.634 |
| 519 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.545 |
-0.789 |
| 520 |
257759_at |
AT3G23070
|
ATCFM3A, CFM3A, CRM family member 3A |
-0.544 |
-0.524 |
| 521 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.544 |
-0.395 |
| 522 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.543 |
-0.564 |
| 523 |
252100_at |
AT3G51110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.541 |
0.270 |
| 524 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.538 |
0.055 |
| 525 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.537 |
-0.309 |
| 526 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.536 |
0.094 |
| 527 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.534 |
-0.744 |
| 528 |
251306_at |
AT3G61260
|
[Remorin family protein] |
-0.534 |
-0.193 |
| 529 |
249290_at |
AT5G41060
|
[DHHC-type zinc finger family protein] |
-0.534 |
-0.260 |
| 530 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.534 |
-0.393 |
| 531 |
262598_at |
AT1G15260
|
unknown |
-0.534 |
-1.107 |
| 532 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.534 |
0.124 |
| 533 |
260166_at |
AT1G79840
|
GL2, GLABRA 2 |
-0.534 |
0.099 |
| 534 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
-0.533 |
-0.003 |
| 535 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.532 |
-0.418 |
| 536 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.531 |
0.410 |
| 537 |
248224_at |
AT5G53490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.531 |
-0.558 |
| 538 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.531 |
-0.665 |
| 539 |
246378_at |
AT1G57620
|
CYB, CYTOPLASMIC BODIES |
-0.530 |
0.156 |
| 540 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.530 |
-0.871 |
| 541 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
-0.529 |
-0.193 |
| 542 |
259012_at |
AT3G07360
|
ATPUB9, ARABIDOPSIS THALIANA PLANT U-BOX 9, PUB9, plant U-box 9 |
-0.529 |
-0.188 |
| 543 |
247425_at |
AT5G62550
|
unknown |
-0.529 |
-0.122 |
| 544 |
259207_at |
AT3G09050
|
unknown |
-0.528 |
-0.639 |
| 545 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
-0.528 |
0.238 |
| 546 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.527 |
-0.266 |
| 547 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
-0.525 |
0.110 |
| 548 |
245041_at |
AT2G26530
|
AR781 |
-0.524 |
0.467 |
| 549 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.523 |
-1.058 |
| 550 |
251641_at |
AT3G57470
|
[Insulinase (Peptidase family M16) family protein] |
-0.521 |
-0.069 |
| 551 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
-0.521 |
-0.588 |
| 552 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.521 |
-0.158 |
| 553 |
251887_at |
AT3G54170
|
ATFIP37, FKBP12 interacting protein 37, FIP37, FKBP12 interacting protein 37 |
-0.521 |
0.149 |
| 554 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.521 |
0.063 |
| 555 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.521 |
-0.338 |
| 556 |
256799_at |
AT3G18560
|
unknown |
-0.520 |
0.445 |
| 557 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.520 |
-0.049 |
| 558 |
255690_at |
AT4G00360
|
ATT1, ABERRANT INDUCTION OF TYPE THREE 1, CYP86A2, cytochrome P450, family 86, subfamily A, polypeptide 2 |
-0.519 |
-0.536 |
| 559 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.518 |
0.170 |
| 560 |
258608_at |
AT3G03020
|
unknown |
-0.517 |
0.001 |
| 561 |
260939_at |
AT1G45180
|
[RING/U-box superfamily protein] |
-0.517 |
0.166 |
| 562 |
250778_at |
AT5G05500
|
MOP10 |
-0.516 |
0.092 |
| 563 |
248540_at |
AT5G50160
|
ATFRO8, FERRIC REDUCTION OXIDASE 8, FRO8, ferric reduction oxidase 8 |
-0.516 |
0.106 |
| 564 |
261455_at |
AT1G21070
|
[Nucleotide-sugar transporter family protein] |
-0.515 |
0.202 |
| 565 |
247597_at |
AT5G60860
|
AtRABA1f, RAB GTPase homolog A1F, RABA1f, RAB GTPase homolog A1F |
-0.515 |
-0.005 |
| 566 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.513 |
-0.725 |
| 567 |
252280_at |
AT3G49260
|
iqd21, IQ-domain 21 |
-0.512 |
-0.643 |
| 568 |
258797_at |
AT3G04730
|
IAA16, indoleacetic acid-induced protein 16 |
-0.512 |
-0.141 |
| 569 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.512 |
0.068 |
| 570 |
257085_at |
AT3G20630
|
ATUBP14, PER1, phosphate deficiency root hair defective1, TTN6, TITAN6, UBP14, ubiquitin-specific protease 14 |
-0.512 |
0.320 |
| 571 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.511 |
0.017 |
| 572 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.511 |
0.126 |
| 573 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
-0.510 |
-0.054 |
| 574 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.509 |
-0.059 |
| 575 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.508 |
0.346 |
| 576 |
251058_at |
AT5G01790
|
unknown |
-0.508 |
-0.951 |
| 577 |
258017_at |
AT3G19370
|
unknown |
-0.508 |
-0.207 |
| 578 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.508 |
0.034 |
| 579 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
-0.507 |
-0.104 |
| 580 |
260819_at |
AT1G06850
|
AtbZIP52, basic leucine-zipper 52, bZIP52, basic leucine-zipper 52 |
-0.506 |
-0.119 |
| 581 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.506 |
-0.943 |
| 582 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.506 |
-0.825 |
| 583 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.506 |
-0.042 |
| 584 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.505 |
-0.636 |
| 585 |
265103_at |
AT1G31070
|
GlcNAc1pUT1, N-acetylglucosamine-1-phosphate uridylyltransferase 1 |
-0.505 |
-0.405 |
| 586 |
262286_at |
AT1G68585
|
unknown |
-0.505 |
-0.415 |
| 587 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.505 |
-0.151 |
| 588 |
254982_at |
AT4G10470
|
unknown |
-0.505 |
-0.606 |
| 589 |
259874_at |
AT1G76660
|
unknown |
-0.505 |
0.055 |
| 590 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.504 |
-0.756 |
| 591 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.504 |
-0.563 |
| 592 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.503 |
-0.624 |
| 593 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.502 |
-1.442 |
| 594 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.501 |
-0.042 |
| 595 |
245690_at |
AT5G04230
|
ATPAL3, PAL3, phenyl alanine ammonia-lyase 3 |
-0.499 |
-0.498 |
| 596 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.499 |
-0.441 |
| 597 |
267511_at |
AT2G45670
|
[calcineurin B subunit-related] |
-0.497 |
0.000 |
| 598 |
267557_at |
AT2G32710
|
ACK2, CYCLIN-DEPENDENT KINASE INHIBITOR 2, ICK7, INTERACTORS OF CDC2 KINASE 7, KRP4, KRP4, KIP-RELATED PROTEIN 4 |
-0.497 |
0.024 |
| 599 |
245759_at |
AT1G66900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.497 |
0.072 |
| 600 |
251968_at |
AT3G53100
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.497 |
-0.349 |