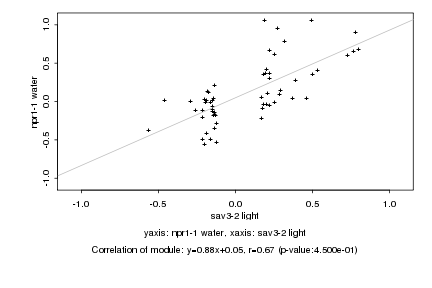
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
sav3-2 light |
Log2 signal ratio
npr1-1 water |
| 1 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.794 |
0.681 |
| 2 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.777 |
0.906 |
| 3 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.765 |
0.661 |
| 4 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.727 |
0.612 |
| 5 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.530 |
0.407 |
| 6 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.494 |
0.355 |
| 7 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
0.491 |
1.063 |
| 8 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.457 |
0.047 |
| 9 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.389 |
0.276 |
| 10 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.370 |
0.048 |
| 11 |
261221_at |
AT1G19960
|
unknown |
0.314 |
0.793 |
| 12 |
245074_at |
AT2G23200
|
[Protein kinase superfamily protein] |
0.289 |
0.144 |
| 13 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.281 |
0.104 |
| 14 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.268 |
0.960 |
| 15 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.253 |
0.616 |
| 16 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.250 |
-0.006 |
| 17 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
0.221 |
0.303 |
| 18 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.221 |
0.670 |
| 19 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
0.217 |
0.371 |
| 20 |
258982_at |
AT3G08870
|
LecRK-VI.1, L-type lectin receptor kinase VI.1 |
0.215 |
-0.045 |
| 21 |
265611_at |
AT2G25510
|
unknown |
0.206 |
0.112 |
| 22 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.200 |
0.422 |
| 23 |
253997_at |
AT4G26090
|
RPS2, RESISTANT TO P. SYRINGAE 2 |
0.199 |
-0.036 |
| 24 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
0.195 |
0.367 |
| 25 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.184 |
1.069 |
| 26 |
253993_at |
AT4G26070
|
ATMEK1, MEK1, MAP kinase/ ERK kinase 1, MKK1, MITOGEN ACTIVATED PROTEIN KINASE KINASE 1, NMAPKK |
0.182 |
0.365 |
| 27 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.182 |
-0.033 |
| 28 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
0.170 |
-0.084 |
| 29 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
0.169 |
0.053 |
| 30 |
257480_at |
AT1G15640
|
unknown |
0.166 |
-0.212 |
| 31 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.567 |
-0.372 |
| 32 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-0.464 |
0.022 |
| 33 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.296 |
0.012 |
| 34 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
-0.265 |
-0.111 |
| 35 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.220 |
-0.491 |
| 36 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.220 |
-0.199 |
| 37 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.215 |
-0.113 |
| 38 |
248801_at |
AT5G47370
|
HAT2 |
-0.207 |
0.034 |
| 39 |
255074_at |
AT4G09100
|
[RING/U-box superfamily protein] |
-0.206 |
-0.560 |
| 40 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.195 |
-0.008 |
| 41 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.191 |
-0.407 |
| 42 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.188 |
0.021 |
| 43 |
253246_at |
AT4G34600
|
unknown |
-0.182 |
0.143 |
| 44 |
250203_at |
AT5G13980
|
[Glycosyl hydrolase family 38 protein] |
-0.176 |
0.126 |
| 45 |
258445_at |
AT3G22400
|
ATLOX5, Arabidopsis thaliana lipoxygenase 5, LOX5 |
-0.168 |
-0.005 |
| 46 |
248969_at |
AT5G45310
|
unknown |
-0.166 |
-0.495 |
| 47 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.154 |
-0.060 |
| 48 |
250396_at |
AT5G10970
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.153 |
0.021 |
| 49 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.152 |
-0.098 |
| 50 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.150 |
-0.126 |
| 51 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.146 |
-0.179 |
| 52 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.146 |
0.045 |
| 53 |
247913_at |
AT5G57510
|
unknown |
-0.142 |
-0.343 |
| 54 |
245944_at |
AT5G19520
|
ATMSL9, MSL9, mechanosensitive channel of small conductance-like 9 |
-0.139 |
-0.140 |
| 55 |
252751_at |
AT3G43430
|
[RING/U-box superfamily protein] |
-0.137 |
0.221 |
| 56 |
264835_at |
AT1G03550
|
AtSCAMP4, SCAMP4, Secretory carrier membrane protein 4 |
-0.137 |
-0.166 |
| 57 |
265246_at |
AT2G43050
|
ATPMEPCRD |
-0.133 |
-0.170 |
| 58 |
250653_at |
AT5G06930
|
[LOCATED IN: chloroplast] |
-0.127 |
-0.285 |
| 59 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.126 |
-0.530 |