|
probeID |
AGICode |
Annotation |
Log2 signal ratio
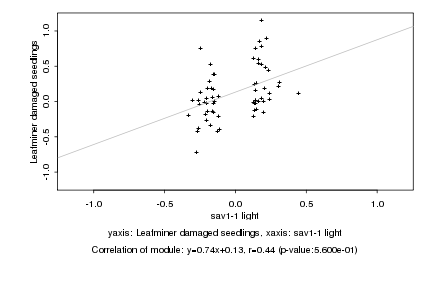
sav1-1 light |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.441 |
0.118 |
| 2 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.305 |
0.270 |
| 3 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.303 |
0.225 |
| 4 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.235 |
0.115 |
| 5 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.234 |
0.030 |
| 6 |
264774_at |
AT1G22890
|
unknown |
0.229 |
0.444 |
| 7 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.216 |
0.903 |
| 8 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.206 |
0.486 |
| 9 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.205 |
0.187 |
| 10 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
0.194 |
0.009 |
| 11 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
0.192 |
-0.143 |
| 12 |
263182_at |
AT1G05575
|
unknown |
0.183 |
1.162 |
| 13 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.180 |
0.792 |
| 14 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
0.177 |
0.535 |
| 15 |
264505_at |
AT1G09380
|
UMAMIT25, Usually multiple acids move in and out Transporters 25 |
0.177 |
0.043 |
| 16 |
266658_at |
AT2G25735
|
unknown |
0.167 |
0.857 |
| 17 |
253315_at |
AT4G33900
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.159 |
0.005 |
| 18 |
249454_at |
AT5G39520
|
unknown |
0.159 |
0.603 |
| 19 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.157 |
0.543 |
| 20 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
0.147 |
0.256 |
| 21 |
265155_at |
AT1G30990
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.146 |
-0.110 |
| 22 |
258078_at |
AT3G25870
|
unknown |
0.145 |
0.257 |
| 23 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.140 |
0.025 |
| 24 |
256442_at |
AT3G10930
|
unknown |
0.139 |
0.757 |
| 25 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
0.137 |
0.157 |
| 26 |
256898_at |
AT3G24650
|
ABI3, ABA INSENSITIVE 3, ABI3, ABSCISIC ACID INSENSITIVE 3, SIS10, SUGAR INSENSITIVE 10 |
0.137 |
-0.016 |
| 27 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.135 |
0.015 |
| 28 |
253439_at |
AT4G32540
|
YUC, YUCCA, YUC1, YUCCA 1 |
0.133 |
-0.019 |
| 29 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
0.133 |
0.245 |
| 30 |
257551_at |
AT3G13270
|
unknown |
0.132 |
-0.115 |
| 31 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.124 |
0.616 |
| 32 |
264860_at |
AT1G24290
|
[AAA-type ATPase family protein] |
0.123 |
-0.200 |
| 33 |
248788_at |
AT5G47430
|
[DWNN domain, a CCHC-type zinc finger] |
0.121 |
-0.001 |
| 34 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.337 |
-0.187 |
| 35 |
266654_at |
AT2G25890
|
[Oleosin family protein] |
-0.306 |
0.016 |
| 36 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
-0.280 |
-0.716 |
| 37 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.272 |
-0.416 |
| 38 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
-0.263 |
0.020 |
| 39 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.261 |
-0.381 |
| 40 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
-0.255 |
-0.035 |
| 41 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.252 |
0.758 |
| 42 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
-0.247 |
0.140 |
| 43 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.223 |
-0.007 |
| 44 |
245326_at |
AT4G14100
|
[transferases, transferring glycosyl groups] |
-0.212 |
-0.175 |
| 45 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.210 |
0.047 |
| 46 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
-0.209 |
-0.263 |
| 47 |
260939_at |
AT1G45180
|
[RING/U-box superfamily protein] |
-0.206 |
-0.021 |
| 48 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.201 |
-0.129 |
| 49 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
-0.200 |
0.193 |
| 50 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.190 |
0.288 |
| 51 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.179 |
-0.327 |
| 52 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.177 |
0.538 |
| 53 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.172 |
0.189 |
| 54 |
245156_at |
AT5G12480
|
CPK7, calmodulin-domain protein kinase 7 |
-0.165 |
0.062 |
| 55 |
250823_at |
AT5G05180
|
unknown |
-0.162 |
-0.140 |
| 56 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.161 |
-0.024 |
| 57 |
263120_at |
AT1G78490
|
CYP708A3, cytochrome P450, family 708, subfamily A, polypeptide 3 |
-0.159 |
0.183 |
| 58 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.156 |
-0.154 |
| 59 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
-0.156 |
0.383 |
| 60 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.154 |
0.395 |
| 61 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
-0.149 |
0.011 |
| 62 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.129 |
-0.418 |
| 63 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.125 |
-0.203 |
| 64 |
265246_at |
AT2G43050
|
ATPMEPCRD |
-0.121 |
0.074 |
| 65 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.119 |
-0.384 |