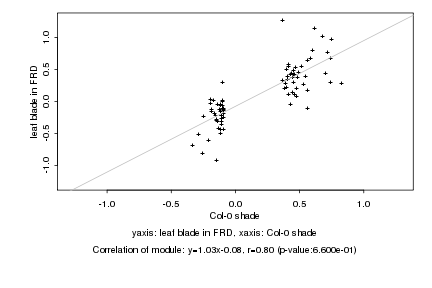
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 shade |
Log2 signal ratio
leaf blade in FRD |
| 1 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.824 |
0.283 |
| 2 |
264323_at |
AT1G04180
|
YUC9, YUCCA 9 |
0.747 |
0.986 |
| 3 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.737 |
0.683 |
| 4 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
0.734 |
0.308 |
| 5 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
0.713 |
0.776 |
| 6 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.701 |
0.447 |
| 7 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.675 |
1.021 |
| 8 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.611 |
1.149 |
| 9 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.597 |
0.799 |
| 10 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
0.579 |
0.679 |
| 11 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.558 |
0.657 |
| 12 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.555 |
0.183 |
| 13 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.555 |
-0.094 |
| 14 |
266814_at |
AT2G44910
|
ATHB-4, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 4, ATHB4, homeobox-leucine zipper protein 4, HB4, homeobox-leucine zipper protein 4 |
0.542 |
0.401 |
| 15 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.529 |
0.272 |
| 16 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.510 |
0.559 |
| 17 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.486 |
0.456 |
| 18 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
0.479 |
0.390 |
| 19 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
0.470 |
0.211 |
| 20 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
0.469 |
0.090 |
| 21 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.464 |
0.541 |
| 22 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.460 |
0.428 |
| 23 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.454 |
0.116 |
| 24 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.446 |
0.432 |
| 25 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.445 |
0.305 |
| 26 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
0.445 |
0.492 |
| 27 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.442 |
0.379 |
| 28 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.438 |
0.145 |
| 29 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.429 |
0.443 |
| 30 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.427 |
-0.031 |
| 31 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.425 |
0.436 |
| 32 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
0.412 |
0.586 |
| 33 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.411 |
0.119 |
| 34 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.410 |
0.553 |
| 35 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.404 |
0.352 |
| 36 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
0.398 |
0.394 |
| 37 |
252204_at |
AT3G50340
|
unknown |
0.394 |
0.221 |
| 38 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.392 |
0.503 |
| 39 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
0.387 |
0.288 |
| 40 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.380 |
0.213 |
| 41 |
248801_at |
AT5G47370
|
HAT2 |
0.363 |
0.340 |
| 42 |
245076_at |
AT2G23170
|
GH3.3 |
0.360 |
1.283 |
| 43 |
252661_at |
AT3G44450
|
unknown |
-0.340 |
-0.677 |
| 44 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
-0.290 |
-0.502 |
| 45 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.264 |
-0.805 |
| 46 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
-0.252 |
-0.227 |
| 47 |
252010_at |
AT3G52740
|
unknown |
-0.211 |
-0.598 |
| 48 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.196 |
0.046 |
| 49 |
260081_at |
AT1G78170
|
unknown |
-0.195 |
-0.023 |
| 50 |
245758_at |
AT1G32240
|
KAN2, KANADI 2 |
-0.188 |
-0.119 |
| 51 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.187 |
-0.147 |
| 52 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.173 |
0.018 |
| 53 |
258919_at |
AT3G10525
|
LGO, LOSS OF GIANT CELLS FROM ORGANS, SMR1, SIAMESE RELATED 1 |
-0.167 |
-0.179 |
| 54 |
252619_at |
AT3G45210
|
unknown |
-0.158 |
-0.208 |
| 55 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.153 |
-0.910 |
| 56 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.153 |
-0.296 |
| 57 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
-0.150 |
-0.270 |
| 58 |
262170_at |
AT1G74940
|
unknown |
-0.147 |
-0.041 |
| 59 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.145 |
-0.300 |
| 60 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.139 |
-0.419 |
| 61 |
257260_at |
AT3G22104
|
[Phototropic-responsive NPH3 family protein] |
-0.132 |
-0.119 |
| 62 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.122 |
-0.427 |
| 63 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
-0.122 |
-0.151 |
| 64 |
253084_at |
AT4G36260
|
SRS2, SHI RELATED SEQUENCE 2, STY2, STYLISH 2 |
-0.122 |
-0.057 |
| 65 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.121 |
-0.494 |
| 66 |
267305_at |
AT2G30070
|
ATKT1, potassium transporter 1, ATKT1P, ATKUP1, KT1, potassium transporter 1, KUP1, POTASSIUM UPTAKE TRANSPORTER 1 |
-0.120 |
-0.040 |
| 67 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.115 |
-0.215 |
| 68 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.115 |
-0.215 |
| 69 |
265309_at |
AT2G20290
|
ATXIG, MYOSIN XI G, XIG, myosin-like protein XIG |
-0.115 |
-0.346 |
| 70 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.114 |
-0.253 |
| 71 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.114 |
-0.304 |
| 72 |
257719_at |
AT3G18440
|
ALMT9, aluminum-activated malate transporter 9, AtALMT9, aluminum-activated malate transporter 9 |
-0.106 |
-0.106 |
| 73 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
-0.105 |
-0.098 |
| 74 |
262827_at |
AT1G13100
|
CYP71B29, cytochrome P450, family 71, subfamily B, polypeptide 29 |
-0.105 |
0.022 |
| 75 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
-0.103 |
0.006 |
| 76 |
258935_at |
AT3G10120
|
unknown |
-0.102 |
0.312 |
| 77 |
259656_at |
AT1G55200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.101 |
-0.060 |
| 78 |
267165_at |
AT2G37710
|
LecRK-IV.1, L-type lectin receptor kinase IV.1, RLK, receptor lectin kinase |
-0.100 |
-0.172 |
| 79 |
262598_at |
AT1G15260
|
unknown |
-0.099 |
-0.125 |
| 80 |
261139_at |
AT1G19700
|
BEL10, BEL1-like homeodomain 10, BLH10, BEL1-LIKE HOMEODOMAIN 10 |
-0.098 |
-0.098 |
| 81 |
249551_at |
AT5G38220
|
[alpha/beta-Hydrolases superfamily protein] |
-0.097 |
-0.115 |
| 82 |
255502_at |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
-0.097 |
-0.244 |
| 83 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
-0.097 |
-0.425 |