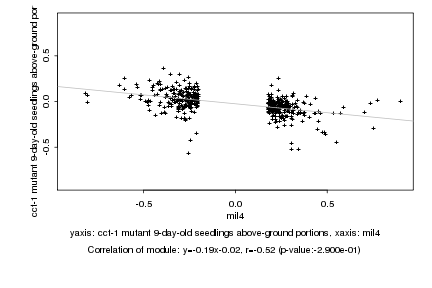
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
mil4 |
Log2 signal ratio
cct-1 mutant 9-day-old seedlings above-ground portions |
| 1 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.897 |
0.003 |
| 2 |
267345_at |
AT2G44240
|
unknown |
0.773 |
0.019 |
| 3 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.748 |
-0.293 |
| 4 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.735 |
-0.013 |
| 5 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.699 |
-0.110 |
| 6 |
253322_at |
AT4G33980
|
unknown |
0.585 |
-0.064 |
| 7 |
261658_at |
AT1G50040
|
unknown |
0.567 |
-0.129 |
| 8 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.546 |
-0.443 |
| 9 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.533 |
-0.124 |
| 10 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.488 |
-0.355 |
| 11 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.480 |
-0.331 |
| 12 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.472 |
-0.333 |
| 13 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.461 |
-0.126 |
| 14 |
253827_at |
AT4G28085
|
unknown |
0.450 |
-0.100 |
| 15 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.450 |
-0.212 |
| 16 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.446 |
-0.302 |
| 17 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.433 |
0.041 |
| 18 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.432 |
-0.121 |
| 19 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
0.429 |
-0.124 |
| 20 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.408 |
-0.030 |
| 21 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.403 |
-0.174 |
| 22 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.385 |
0.060 |
| 23 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
0.376 |
-0.143 |
| 24 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.372 |
-0.115 |
| 25 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.372 |
-0.014 |
| 26 |
261822_at |
AT1G11380
|
[PLAC8 family protein] |
0.370 |
-0.005 |
| 27 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.367 |
-0.128 |
| 28 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.363 |
-0.208 |
| 29 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.353 |
-0.131 |
| 30 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.351 |
-0.110 |
| 31 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
0.349 |
-0.097 |
| 32 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
0.341 |
-0.111 |
| 33 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.340 |
-0.517 |
| 34 |
261851_at |
AT1G50460
|
ATHKL1, HKL1, hexokinase-like 1 |
0.335 |
-0.132 |
| 35 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
0.333 |
-0.059 |
| 36 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.333 |
0.020 |
| 37 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
0.329 |
-0.044 |
| 38 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.325 |
-0.015 |
| 39 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.320 |
-0.061 |
| 40 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.314 |
0.097 |
| 41 |
248889_at |
AT5G46230
|
unknown |
0.313 |
-0.123 |
| 42 |
253811_at |
AT4G28190
|
ULT, ULTRAPETALA, ULT1, ULTRAPETALA1 |
0.313 |
-0.093 |
| 43 |
261117_at |
AT1G75310
|
AUL1, auxilin-like 1 |
0.310 |
-0.092 |
| 44 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.310 |
-0.029 |
| 45 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.310 |
0.063 |
| 46 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.308 |
0.076 |
| 47 |
246926_at |
AT5G25240
|
unknown |
0.308 |
-0.194 |
| 48 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.305 |
-0.161 |
| 49 |
256848_at |
AT3G27960
|
KLCR2, kinesin light chain-related 2 |
0.305 |
-0.114 |
| 50 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.305 |
-0.516 |
| 51 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
0.305 |
-0.183 |
| 52 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.303 |
-0.253 |
| 53 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
0.300 |
-0.451 |
| 54 |
264910_at |
AT1G61100
|
[disease resistance protein (TIR class), putative] |
0.299 |
-0.048 |
| 55 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.294 |
-0.118 |
| 56 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.294 |
-0.112 |
| 57 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.291 |
-0.048 |
| 58 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.289 |
-0.019 |
| 59 |
266424_at |
AT2G41330
|
[Glutaredoxin family protein] |
0.286 |
-0.012 |
| 60 |
253478_at |
AT4G32350
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.286 |
-0.078 |
| 61 |
260688_at |
AT1G17665
|
unknown |
0.286 |
-0.071 |
| 62 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.283 |
-0.034 |
| 63 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.281 |
-0.018 |
| 64 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
0.280 |
0.011 |
| 65 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.279 |
-0.159 |
| 66 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
0.279 |
-0.106 |
| 67 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.279 |
-0.045 |
| 68 |
264822_at |
AT1G03457
|
AtBRN2, BRN2, Bruno-like 2 |
0.277 |
-0.022 |
| 69 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
0.277 |
-0.064 |
| 70 |
252173_at |
AT3G50650
|
[GRAS family transcription factor] |
0.276 |
-0.036 |
| 71 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.275 |
0.060 |
| 72 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
0.275 |
0.005 |
| 73 |
249577_at |
AT5G37710
|
[alpha/beta-Hydrolases superfamily protein] |
0.273 |
-0.017 |
| 74 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
0.273 |
-0.158 |
| 75 |
256513_at |
AT1G31480
|
SGR2, SHOOT GRAVITROPISM 2 |
0.271 |
0.004 |
| 76 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
0.271 |
-0.068 |
| 77 |
249409_at |
AT5G40340
|
[Tudor/PWWP/MBT superfamily protein] |
0.271 |
-0.069 |
| 78 |
264238_at |
AT1G54740
|
unknown |
0.269 |
-0.156 |
| 79 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.268 |
-0.042 |
| 80 |
263118_at |
AT1G03090
|
MCCA |
0.267 |
0.035 |
| 81 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.266 |
-0.260 |
| 82 |
262910_at |
AT1G59710
|
unknown |
0.265 |
-0.063 |
| 83 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.264 |
-0.118 |
| 84 |
259065_at |
AT3G07520
|
ATGLR1.4, GLR1.4, glutamate receptor 1.4 |
0.264 |
-0.094 |
| 85 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.263 |
-0.037 |
| 86 |
265771_at |
AT2G48030
|
[DNAse I-like superfamily protein] |
0.263 |
-0.200 |
| 87 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.261 |
-0.181 |
| 88 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.256 |
-0.007 |
| 89 |
246446_at |
AT5G17640
|
ASG1, Abiotic Stress Gene 1 |
0.256 |
-0.034 |
| 90 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
0.255 |
-0.025 |
| 91 |
246063_at |
AT5G19340
|
unknown |
0.255 |
0.062 |
| 92 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
0.255 |
-0.053 |
| 93 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.254 |
0.008 |
| 94 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.251 |
-0.146 |
| 95 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.251 |
-0.057 |
| 96 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.250 |
-0.182 |
| 97 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.250 |
0.013 |
| 98 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
0.249 |
0.021 |
| 99 |
246222_at |
AT4G36900
|
DEAR4, DREB AND EAR MOTIF PROTEIN 4, RAP2.10, related to AP2 10 |
0.247 |
-0.159 |
| 100 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.246 |
-0.024 |
| 101 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.246 |
-0.165 |
| 102 |
252539_at |
AT3G45730
|
unknown |
0.245 |
-0.055 |
| 103 |
259996_at |
AT1G67910
|
unknown |
0.245 |
-0.024 |
| 104 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.242 |
-0.111 |
| 105 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.241 |
-0.060 |
| 106 |
263222_at |
AT1G30640
|
[Protein kinase family protein] |
0.241 |
-0.016 |
| 107 |
246843_at |
AT5G26740
|
unknown |
0.241 |
0.025 |
| 108 |
263499_at |
AT2G42580
|
TTL3, tetratricopetide-repeat thioredoxin-like 3, VIT, VHI-INTERACTING TPR CONTAINING PROTEIN |
0.240 |
-0.121 |
| 109 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.239 |
-0.212 |
| 110 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.239 |
-0.211 |
| 111 |
266163_at |
AT2G28130
|
unknown |
0.238 |
-0.083 |
| 112 |
255931_at |
AT1G12710
|
AtPP2-A12, phloem protein 2-A12, PP2-A12, phloem protein 2-A12 |
0.236 |
-0.053 |
| 113 |
261727_at |
AT1G76090
|
SMT3, sterol methyltransferase 3 |
0.236 |
-0.154 |
| 114 |
249763_at |
AT5G24010
|
[Protein kinase superfamily protein] |
0.236 |
0.019 |
| 115 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
0.236 |
-0.112 |
| 116 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.236 |
-0.021 |
| 117 |
258225_at |
AT3G15630
|
unknown |
0.235 |
-0.069 |
| 118 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
0.234 |
-0.041 |
| 119 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
0.234 |
0.031 |
| 120 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.232 |
-0.005 |
| 121 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.231 |
0.130 |
| 122 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
0.231 |
-0.120 |
| 123 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.230 |
0.258 |
| 124 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
0.229 |
-0.071 |
| 125 |
258855_at |
AT3G02070
|
[Cysteine proteinases superfamily protein] |
0.229 |
-0.073 |
| 126 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
0.228 |
-0.102 |
| 127 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.227 |
-0.088 |
| 128 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.226 |
-0.051 |
| 129 |
267397_at |
AT1G76170
|
[2-thiocytidine tRNA biosynthesis protein, TtcA] |
0.226 |
0.044 |
| 130 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.224 |
0.008 |
| 131 |
260744_at |
AT1G15010
|
unknown |
0.224 |
-0.281 |
| 132 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.224 |
-0.090 |
| 133 |
247394_at |
AT5G62865
|
unknown |
0.223 |
-0.100 |
| 134 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.221 |
-0.218 |
| 135 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.221 |
-0.113 |
| 136 |
260686_at |
AT1G17620
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.220 |
-0.099 |
| 137 |
266956_at |
AT2G34510
|
unknown |
0.219 |
-0.019 |
| 138 |
260367_at |
AT1G69760
|
unknown |
0.218 |
-0.188 |
| 139 |
258682_at |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
0.218 |
-0.081 |
| 140 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.217 |
-0.012 |
| 141 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.217 |
-0.156 |
| 142 |
260668_at |
AT1G19530
|
unknown |
0.215 |
-0.157 |
| 143 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
0.215 |
-0.226 |
| 144 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.214 |
-0.025 |
| 145 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
0.213 |
-0.126 |
| 146 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.211 |
-0.069 |
| 147 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.211 |
-0.017 |
| 148 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.211 |
-0.050 |
| 149 |
248796_at |
AT5G47180
|
[Plant VAMP (vesicle-associated membrane protein) family protein] |
0.208 |
-0.092 |
| 150 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.208 |
-0.034 |
| 151 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.208 |
-0.062 |
| 152 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.207 |
-0.082 |
| 153 |
264770_at |
AT1G23030
|
[ARM repeat superfamily protein] |
0.206 |
-0.007 |
| 154 |
254605_at |
AT4G18950
|
[Integrin-linked protein kinase family] |
0.206 |
-0.032 |
| 155 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
0.205 |
-0.091 |
| 156 |
254043_at |
AT4G25990
|
CIL |
0.204 |
-0.143 |
| 157 |
246917_at |
AT5G25280
|
[serine-rich protein-related] |
0.204 |
0.003 |
| 158 |
259530_at |
AT1G12450
|
[SNARE associated Golgi protein family] |
0.203 |
-0.050 |
| 159 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
0.202 |
-0.092 |
| 160 |
262220_at |
AT1G74740
|
ATCPK30, CDPK1A, CALCIUM-DEPENDENT PROTEIN KINASE 1A, CPK30, calcium-dependent protein kinase 30 |
0.202 |
0.013 |
| 161 |
250936_at |
AT5G03120
|
unknown |
0.201 |
-0.191 |
| 162 |
267209_at |
AT2G30930
|
unknown |
0.201 |
-0.156 |
| 163 |
246995_at |
AT5G67470
|
ATFH6, ARABIDOPSIS FORMIN HOMOLOG 6, FH6, formin homolog 6 |
0.201 |
-0.033 |
| 164 |
259103_at |
AT3G11690
|
unknown |
0.199 |
-0.114 |
| 165 |
245906_at |
AT5G11070
|
unknown |
0.198 |
-0.002 |
| 166 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
0.198 |
-0.021 |
| 167 |
265938_at |
AT2G19620
|
NDL3, N-MYC downregulated-like 3 |
0.197 |
-0.076 |
| 168 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.197 |
-0.098 |
| 169 |
267318_at |
AT2G34770
|
ATFAH1, ARABIDOPSIS FATTY ACID HYDROXYLASE 1, FAH1, fatty acid hydroxylase 1 |
0.196 |
-0.040 |
| 170 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
0.196 |
-0.101 |
| 171 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.196 |
0.054 |
| 172 |
264517_at |
AT1G10120
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.196 |
-0.040 |
| 173 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.195 |
0.078 |
| 174 |
253647_at |
AT4G29950
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.195 |
-0.015 |
| 175 |
246796_at |
AT5G26770
|
unknown |
0.195 |
-0.040 |
| 176 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
0.194 |
0.011 |
| 177 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.193 |
0.184 |
| 178 |
247149_at |
AT5G65660
|
[hydroxyproline-rich glycoprotein family protein] |
0.192 |
-0.106 |
| 179 |
251406_at |
AT3G60260
|
[ELMO/CED-12 family protein] |
0.192 |
-0.063 |
| 180 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
0.191 |
-0.034 |
| 181 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.191 |
-0.064 |
| 182 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.189 |
-0.088 |
| 183 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
0.189 |
-0.088 |
| 184 |
251105_at |
AT5G01730
|
ATSCAR4, SCAR4, SCAR family protein 4, WAVE3 |
0.189 |
-0.033 |
| 185 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
0.189 |
0.017 |
| 186 |
249580_at |
AT5G37740
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.189 |
-0.066 |
| 187 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
0.188 |
0.039 |
| 188 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
0.187 |
0.031 |
| 189 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
0.187 |
-0.078 |
| 190 |
262491_at |
AT1G21650
|
SECA2 |
0.186 |
0.008 |
| 191 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.183 |
-0.025 |
| 192 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
0.183 |
-0.121 |
| 193 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.183 |
-0.240 |
| 194 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.183 |
0.065 |
| 195 |
253543_at |
AT4G31270
|
[sequence-specific DNA binding transcription factors] |
0.182 |
-0.019 |
| 196 |
260734_at |
AT1G17600
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.181 |
-0.066 |
| 197 |
259106_at |
AT3G05490
|
RALFL22, ralf-like 22 |
0.180 |
-0.108 |
| 198 |
263854_at |
AT2G04430
|
atnudt5, nudix hydrolase homolog 5, NUDT5, nudix hydrolase homolog 5 |
0.180 |
0.024 |
| 199 |
259232_at |
AT3G11420
|
unknown |
0.180 |
-0.081 |
| 200 |
255061_at |
AT4G08930
|
APRL6, APR-like 6, ATAPRL6, APR-like 6 |
0.179 |
0.085 |
| 201 |
266780_at |
AT2G29110
|
ATGLR2.8, glutamate receptor 2.8, GLR2.8, GLR2.8, glutamate receptor 2.8 |
0.179 |
0.000 |
| 202 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.179 |
-0.063 |
| 203 |
247944_at |
AT5G57100
|
[Nucleotide/sugar transporter family protein] |
0.179 |
-0.020 |
| 204 |
266732_at |
AT2G03240
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.178 |
-0.061 |
| 205 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
0.177 |
-0.083 |
| 206 |
257990_at |
AT3G19860
|
bHLH121, basic Helix-Loop-Helix 121 |
0.177 |
-0.074 |
| 207 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.177 |
-0.011 |
| 208 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
0.177 |
-0.018 |
| 209 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
0.176 |
-0.120 |
| 210 |
249069_at |
AT5G44010
|
unknown |
0.176 |
-0.021 |
| 211 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.176 |
-0.063 |
| 212 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.818 |
0.089 |
| 213 |
265892_at |
AT2G15020
|
unknown |
-0.810 |
0.069 |
| 214 |
262211_at |
AT1G74930
|
ORA47 |
-0.810 |
-0.007 |
| 215 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.634 |
0.185 |
| 216 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.609 |
0.261 |
| 217 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.606 |
0.140 |
| 218 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-0.580 |
0.052 |
| 219 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
-0.570 |
0.066 |
| 220 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.542 |
0.194 |
| 221 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.536 |
0.161 |
| 222 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.522 |
0.024 |
| 223 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-0.522 |
0.069 |
| 224 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.513 |
0.069 |
| 225 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.493 |
0.007 |
| 226 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
-0.483 |
-0.000 |
| 227 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
-0.478 |
-0.035 |
| 228 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
-0.476 |
0.015 |
| 229 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.473 |
0.235 |
| 230 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.473 |
-0.095 |
| 231 |
257925_at |
AT3G23170
|
unknown |
-0.463 |
0.007 |
| 232 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.455 |
0.162 |
| 233 |
253643_at |
AT4G29780
|
unknown |
-0.453 |
0.126 |
| 234 |
253859_at |
AT4G27657
|
unknown |
-0.447 |
0.186 |
| 235 |
263384_at |
AT2G40130
|
SMXL8, SMAX1-like 8 |
-0.442 |
0.076 |
| 236 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.440 |
-0.145 |
| 237 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.429 |
0.079 |
| 238 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
-0.425 |
0.204 |
| 239 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.423 |
0.189 |
| 240 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.416 |
-0.039 |
| 241 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.414 |
0.223 |
| 242 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.412 |
-0.003 |
| 243 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-0.412 |
0.135 |
| 244 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.411 |
0.193 |
| 245 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.411 |
0.129 |
| 246 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.407 |
-0.124 |
| 247 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
-0.398 |
0.137 |
| 248 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.397 |
0.371 |
| 249 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.390 |
-0.059 |
| 250 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
-0.390 |
-0.111 |
| 251 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.386 |
0.144 |
| 252 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
-0.384 |
-0.129 |
| 253 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.382 |
0.054 |
| 254 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.380 |
0.086 |
| 255 |
255502_at |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
-0.374 |
0.158 |
| 256 |
260602_at |
AT1G55920
|
ATSERAT2;1, serine acetyltransferase 2;1, SAT1, SERINE ACETYLTRANSFERASE 1, SAT5, SERINE ACETYLTRANSFERASE 5, SERAT2;1, serine acetyltransferase 2;1 |
-0.373 |
0.058 |
| 257 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
-0.368 |
-0.053 |
| 258 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.365 |
-0.005 |
| 259 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.361 |
-0.015 |
| 260 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
-0.358 |
0.301 |
| 261 |
247753_at |
AT5G59070
|
[UDP-Glycosyltransferase superfamily protein] |
-0.357 |
0.043 |
| 262 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.354 |
0.127 |
| 263 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.353 |
0.125 |
| 264 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.353 |
-0.051 |
| 265 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
-0.352 |
-0.031 |
| 266 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-0.347 |
0.049 |
| 267 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.347 |
0.022 |
| 268 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.346 |
0.168 |
| 269 |
251058_at |
AT5G01790
|
unknown |
-0.342 |
0.140 |
| 270 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
-0.339 |
0.095 |
| 271 |
265184_at |
AT1G23710
|
unknown |
-0.334 |
-0.009 |
| 272 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
-0.333 |
0.023 |
| 273 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.332 |
0.088 |
| 274 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.332 |
0.139 |
| 275 |
256185_at |
AT1G51700
|
ADOF1, DOF zinc finger protein 1, DOF1, DOF zinc finger protein 1 |
-0.331 |
-0.008 |
| 276 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
-0.331 |
0.062 |
| 277 |
246997_at |
AT5G67390
|
unknown |
-0.326 |
0.002 |
| 278 |
261772_at |
AT1G76240
|
unknown |
-0.326 |
-0.165 |
| 279 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
-0.323 |
0.142 |
| 280 |
254145_at |
AT4G24700
|
unknown |
-0.323 |
0.214 |
| 281 |
257057_at |
AT3G15310
|
unknown |
-0.321 |
0.007 |
| 282 |
256891_at |
AT3G19030
|
unknown |
-0.315 |
0.036 |
| 283 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.315 |
-0.106 |
| 284 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.314 |
-0.099 |
| 285 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.314 |
-0.044 |
| 286 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.313 |
0.081 |
| 287 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.313 |
-0.077 |
| 288 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.312 |
0.030 |
| 289 |
249939_at |
AT5G22430
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.312 |
-0.067 |
| 290 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
-0.312 |
-0.061 |
| 291 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-0.310 |
0.120 |
| 292 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.309 |
0.176 |
| 293 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.307 |
-0.015 |
| 294 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.306 |
0.305 |
| 295 |
261315_at |
AT1G53170
|
ATERF-8, ATERF8, ETHYLENE RESPONSE ELEMENT BINDING FACTOR 4, ERF8, ethylene response factor 8 |
-0.305 |
0.090 |
| 296 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.305 |
-0.055 |
| 297 |
252010_at |
AT3G52740
|
unknown |
-0.302 |
0.118 |
| 298 |
261758_at |
AT1G08250
|
ADT6, arogenate dehydratase 6, AtADT6, Arabidopsis thaliana arogenate dehydratase 6 |
-0.302 |
0.098 |
| 299 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
-0.301 |
0.017 |
| 300 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.298 |
-0.176 |
| 301 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.295 |
0.131 |
| 302 |
257949_at |
AT3G21750
|
UGT71B1, UDP-glucosyl transferase 71B1 |
-0.295 |
-0.042 |
| 303 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.294 |
0.076 |
| 304 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.292 |
0.008 |
| 305 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
-0.292 |
0.033 |
| 306 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
-0.291 |
-0.084 |
| 307 |
258982_at |
AT3G08870
|
LecRK-VI.1, L-type lectin receptor kinase VI.1 |
-0.291 |
0.072 |
| 308 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.289 |
-0.050 |
| 309 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
-0.288 |
0.104 |
| 310 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.286 |
-0.067 |
| 311 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.284 |
0.064 |
| 312 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.282 |
-0.045 |
| 313 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.281 |
-0.124 |
| 314 |
265620_at |
AT2G27310
|
[F-box family protein] |
-0.280 |
-0.176 |
| 315 |
253830_at |
AT4G27652
|
unknown |
-0.280 |
0.230 |
| 316 |
262811_at |
AT1G11700
|
unknown |
-0.278 |
-0.182 |
| 317 |
248509_at |
AT5G50335
|
unknown |
-0.277 |
-0.195 |
| 318 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.276 |
0.153 |
| 319 |
245041_at |
AT2G26530
|
AR781 |
-0.276 |
-0.200 |
| 320 |
263182_at |
AT1G05575
|
unknown |
-0.275 |
-0.031 |
| 321 |
252346_at |
AT3G48650
|
[pseudogene] |
-0.274 |
-0.048 |
| 322 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.273 |
0.024 |
| 323 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.273 |
0.022 |
| 324 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.272 |
0.146 |
| 325 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
-0.271 |
-0.054 |
| 326 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.271 |
0.141 |
| 327 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.270 |
0.018 |
| 328 |
258275_at |
AT3G15760
|
unknown |
-0.269 |
0.111 |
| 329 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.269 |
-0.040 |
| 330 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.269 |
0.067 |
| 331 |
263931_at |
AT2G36220
|
unknown |
-0.268 |
-0.039 |
| 332 |
267069_at |
AT2G41010
|
ATCAMBP25, calmodulin (CAM)-binding protein of 25 kDa, CAMBP25, calmodulin (CAM)-binding protein of 25 kDa |
-0.268 |
0.031 |
| 333 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
-0.264 |
0.012 |
| 334 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.264 |
0.063 |
| 335 |
246439_at |
AT5G17600
|
[RING/U-box superfamily protein] |
-0.262 |
0.083 |
| 336 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.261 |
0.148 |
| 337 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.261 |
-0.052 |
| 338 |
252619_at |
AT3G45210
|
unknown |
-0.260 |
0.053 |
| 339 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
-0.260 |
0.081 |
| 340 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.258 |
0.101 |
| 341 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.258 |
-0.048 |
| 342 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.258 |
-0.052 |
| 343 |
267623_at |
AT2G39650
|
unknown |
-0.257 |
0.025 |
| 344 |
252661_at |
AT3G44450
|
unknown |
-0.257 |
0.087 |
| 345 |
249614_at |
AT5G37300
|
WSD1 |
-0.257 |
-0.560 |
| 346 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
-0.256 |
-0.008 |
| 347 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.256 |
0.265 |
| 348 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.255 |
-0.177 |
| 349 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.255 |
-0.049 |
| 350 |
247707_at |
AT5G59450
|
[GRAS family transcription factor] |
-0.255 |
-0.020 |
| 351 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.253 |
0.204 |
| 352 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-0.253 |
0.084 |
| 353 |
247751_at |
AT5G59050
|
unknown |
-0.253 |
0.032 |
| 354 |
263545_at |
AT2G21560
|
unknown |
-0.253 |
-0.037 |
| 355 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.252 |
0.168 |
| 356 |
257053_at |
AT3G15210
|
ATERF-4, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4, ATERF4, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4, ERF4, ethylene responsive element binding factor 4, RAP2.5, RELATED TO AP2 5 |
-0.252 |
-0.035 |
| 357 |
255032_at |
AT4G09500
|
[UDP-Glycosyltransferase superfamily protein] |
-0.252 |
-0.036 |
| 358 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
-0.251 |
-0.016 |
| 359 |
254592_at |
AT4G18880
|
AT-HSFA4A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A4A, HSF A4A, heat shock transcription factor A4A |
-0.249 |
0.069 |
| 360 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.248 |
-0.019 |
| 361 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.248 |
-0.421 |
| 362 |
247509_at |
AT5G62020
|
AT-HSFB2A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR B2A, HSFB2A, heat shock transcription factor B2A |
-0.247 |
-0.062 |
| 363 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.247 |
0.077 |
| 364 |
247708_at |
AT5G59550
|
unknown |
-0.246 |
-0.002 |
| 365 |
261193_at |
AT1G32920
|
unknown |
-0.246 |
0.085 |
| 366 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.245 |
-0.069 |
| 367 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
-0.245 |
-0.027 |
| 368 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
-0.244 |
0.056 |
| 369 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.244 |
0.015 |
| 370 |
262236_at |
AT1G48330
|
unknown |
-0.242 |
-0.060 |
| 371 |
248028_at |
AT5G55620
|
unknown |
-0.241 |
-0.102 |
| 372 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.241 |
0.132 |
| 373 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
-0.241 |
0.004 |
| 374 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
-0.240 |
0.143 |
| 375 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
-0.238 |
0.033 |
| 376 |
267166_at |
AT2G37720
|
TBL15, TRICHOME BIREFRINGENCE-LIKE 15 |
-0.236 |
0.053 |
| 377 |
247716_at |
AT5G59350
|
unknown |
-0.235 |
-0.016 |
| 378 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
-0.233 |
0.077 |
| 379 |
266396_at |
AT2G38790
|
unknown |
-0.233 |
0.057 |
| 380 |
264444_at |
AT1G27360
|
SPL11, squamosa promoter-like 11 |
-0.230 |
0.079 |
| 381 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
-0.229 |
0.108 |
| 382 |
254683_at |
AT4G13800
|
unknown |
-0.228 |
-0.030 |
| 383 |
250828_at |
AT5G05250
|
unknown |
-0.228 |
0.140 |
| 384 |
253819_at |
AT4G28350
|
LecRK-VII.2, L-type lectin receptor kinase VII.2 |
-0.227 |
0.012 |
| 385 |
261319_at |
AT1G53090
|
SPA4, SPA1-related 4 |
-0.227 |
0.154 |
| 386 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.227 |
-0.117 |
| 387 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.227 |
0.165 |
| 388 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
-0.225 |
0.001 |
| 389 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.224 |
0.069 |
| 390 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.223 |
0.086 |
| 391 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.222 |
0.039 |
| 392 |
245755_at |
AT1G35210
|
unknown |
-0.221 |
-0.025 |
| 393 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
-0.220 |
0.072 |
| 394 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.220 |
0.084 |
| 395 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.220 |
0.077 |
| 396 |
254178_at |
AT4G23880
|
unknown |
-0.219 |
0.088 |
| 397 |
265742_at |
AT2G01290
|
RPI2, ribose-5-phosphate isomerase 2 |
-0.219 |
0.025 |
| 398 |
261597_at |
AT1G49780
|
PUB26, plant U-box 26 |
-0.219 |
0.052 |
| 399 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.219 |
0.033 |
| 400 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
-0.218 |
0.036 |
| 401 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.217 |
0.084 |
| 402 |
260477_at |
AT1G11050
|
[Protein kinase superfamily protein] |
-0.217 |
-0.063 |
| 403 |
246233_at |
AT4G36550
|
[ARM repeat superfamily protein] |
-0.216 |
0.023 |
| 404 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
-0.215 |
0.020 |
| 405 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.214 |
0.199 |
| 406 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
-0.213 |
-0.348 |
| 407 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
-0.212 |
0.126 |
| 408 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.212 |
-0.037 |
| 409 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.211 |
-0.017 |
| 410 |
264583_at |
AT1G05170
|
[Galactosyltransferase family protein] |
-0.211 |
0.058 |
| 411 |
253535_at |
AT4G31550
|
ATWRKY11, WRKY11, WRKY DNA-binding protein 11 |
-0.211 |
0.011 |
| 412 |
247693_at |
AT5G59730
|
ATEXO70H7, exocyst subunit exo70 family protein H7, EXO70H7, exocyst subunit exo70 family protein H7 |
-0.211 |
-0.048 |
| 413 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.211 |
0.077 |
| 414 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.210 |
-0.013 |
| 415 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.210 |
0.167 |
| 416 |
253580_at |
AT4G30400
|
[RING/U-box superfamily protein] |
-0.208 |
0.043 |
| 417 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.208 |
-0.002 |
| 418 |
266259_at |
AT2G27830
|
unknown |
-0.206 |
0.056 |
| 419 |
256793_at |
AT3G22160
|
JAV1, jasmonate-associated VQ motif gene 1 |
-0.206 |
0.021 |
| 420 |
258038_at |
AT3G21260
|
GLTP3, GLYCOLIPID TRANSFER PROTEIN 3 |
-0.205 |
0.142 |
| 421 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.205 |
0.085 |