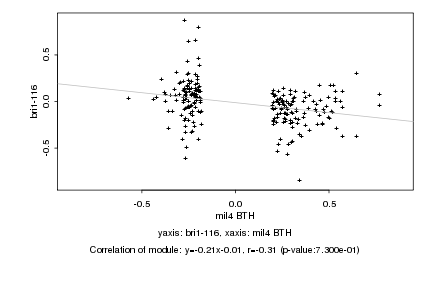
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
mil4 BTH |
Log2 signal ratio
bri1-116 |
| 1 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.766 |
-0.036 |
| 2 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.766 |
0.085 |
| 3 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.643 |
0.311 |
| 4 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.642 |
-0.375 |
| 5 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.571 |
-0.367 |
| 6 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.571 |
0.111 |
| 7 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.569 |
-0.056 |
| 8 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.559 |
0.001 |
| 9 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.535 |
-0.282 |
| 10 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.533 |
0.008 |
| 11 |
259893_at |
AT1G71390
|
AtRLP11, receptor like protein 11, RLP11, receptor like protein 11 |
0.532 |
0.109 |
| 12 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.531 |
0.041 |
| 13 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.522 |
0.182 |
| 14 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.518 |
-0.111 |
| 15 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.511 |
-0.100 |
| 16 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.504 |
0.172 |
| 17 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.502 |
-0.179 |
| 18 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.493 |
-0.171 |
| 19 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.492 |
0.045 |
| 20 |
253322_at |
AT4G33980
|
unknown |
0.484 |
-0.046 |
| 21 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.472 |
-0.116 |
| 22 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.466 |
-0.085 |
| 23 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.465 |
-0.231 |
| 24 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.460 |
-0.239 |
| 25 |
259805_at |
AT1G47890
|
AtRLP7, receptor like protein 7, RLP7, receptor like protein 7 |
0.450 |
0.015 |
| 26 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.449 |
-0.142 |
| 27 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.446 |
0.172 |
| 28 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.434 |
-0.243 |
| 29 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.430 |
-0.024 |
| 30 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.430 |
-0.104 |
| 31 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.425 |
-0.084 |
| 32 |
260117_at |
AT1G33950
|
[Avirulence induced gene (AIG1) family protein] |
0.417 |
0.006 |
| 33 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.393 |
-0.305 |
| 34 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
0.392 |
0.066 |
| 35 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.386 |
-0.072 |
| 36 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.374 |
0.052 |
| 37 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.373 |
-0.257 |
| 38 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
0.369 |
-0.121 |
| 39 |
260688_at |
AT1G17665
|
unknown |
0.368 |
0.098 |
| 40 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.361 |
0.006 |
| 41 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.359 |
-0.165 |
| 42 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.348 |
-0.369 |
| 43 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.340 |
-0.352 |
| 44 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.338 |
-0.843 |
| 45 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.333 |
-0.186 |
| 46 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.331 |
-0.231 |
| 47 |
260668_at |
AT1G19530
|
unknown |
0.326 |
-0.080 |
| 48 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.326 |
-0.107 |
| 49 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.320 |
-0.176 |
| 50 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.318 |
0.113 |
| 51 |
246498_at |
AT5G16230
|
[Plant stearoyl-acyl-carrier-protein desaturase family protein] |
0.312 |
0.051 |
| 52 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.310 |
0.009 |
| 53 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.310 |
0.041 |
| 54 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.304 |
-0.122 |
| 55 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.303 |
-0.223 |
| 56 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.302 |
0.006 |
| 57 |
248708_at |
AT5G48560
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.301 |
-0.275 |
| 58 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.300 |
-0.421 |
| 59 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
0.298 |
-0.431 |
| 60 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.297 |
-0.115 |
| 61 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.297 |
-0.023 |
| 62 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.294 |
-0.201 |
| 63 |
267345_at |
AT2G44240
|
unknown |
0.293 |
0.129 |
| 64 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.292 |
-0.235 |
| 65 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.292 |
0.084 |
| 66 |
267036_at |
AT2G38465
|
unknown |
0.290 |
-0.038 |
| 67 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.283 |
-0.012 |
| 68 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.280 |
-0.451 |
| 69 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.275 |
-0.565 |
| 70 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.275 |
-0.136 |
| 71 |
262085_at |
AT1G56060
|
unknown |
0.273 |
-0.075 |
| 72 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.272 |
0.007 |
| 73 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.272 |
-0.059 |
| 74 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.270 |
-0.131 |
| 75 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.264 |
-0.123 |
| 76 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.264 |
-0.192 |
| 77 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
0.263 |
-0.213 |
| 78 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.262 |
-0.125 |
| 79 |
249057_at |
AT5G44480
|
DUR, DEFECTIVE UGE IN ROOT |
0.261 |
-0.179 |
| 80 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.259 |
-0.063 |
| 81 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
0.258 |
-0.032 |
| 82 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.256 |
-0.023 |
| 83 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.255 |
0.067 |
| 84 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.255 |
0.150 |
| 85 |
258682_at |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
0.254 |
-0.224 |
| 86 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.251 |
-0.010 |
| 87 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.251 |
0.005 |
| 88 |
245076_at |
AT2G23170
|
GH3.3 |
0.249 |
0.019 |
| 89 |
254292_at |
AT4G23030
|
[MATE efflux family protein] |
0.245 |
-0.075 |
| 90 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
0.244 |
-0.068 |
| 91 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.240 |
0.042 |
| 92 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.239 |
-0.402 |
| 93 |
253168_at |
AT4G35070
|
[SBP (S-ribonuclease binding protein) family protein] |
0.237 |
-0.120 |
| 94 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.235 |
-0.030 |
| 95 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.233 |
0.027 |
| 96 |
247177_at |
AT5G65300
|
unknown |
0.228 |
0.112 |
| 97 |
248715_at |
AT5G48290
|
[Heavy metal transport/detoxification superfamily protein ] |
0.227 |
-0.010 |
| 98 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.226 |
-0.452 |
| 99 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.224 |
-0.529 |
| 100 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.222 |
-0.161 |
| 101 |
254245_at |
AT4G23240
|
CRK16, cysteine-rich RLK (RECEPTOR-like protein kinase) 16 |
0.221 |
-0.137 |
| 102 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.220 |
0.028 |
| 103 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.218 |
-0.224 |
| 104 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.217 |
0.006 |
| 105 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.212 |
0.065 |
| 106 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.209 |
-0.072 |
| 107 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.208 |
-0.177 |
| 108 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.207 |
0.058 |
| 109 |
253044_at |
AT4G37290
|
unknown |
0.203 |
-0.242 |
| 110 |
255342_at |
AT4G04510
|
CRK38, cysteine-rich RLK (RECEPTOR-like protein kinase) 38 |
0.202 |
-0.002 |
| 111 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.202 |
-0.195 |
| 112 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
0.201 |
0.084 |
| 113 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.201 |
-0.212 |
| 114 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
0.200 |
0.123 |
| 115 |
266052_at |
AT2G40740
|
ATWRKY55, WRKY DNA-BINDING PROTEIN 55, WRKY55, WRKY DNA-binding protein 55 |
0.199 |
-0.076 |
| 116 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
0.198 |
-0.068 |
| 117 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.196 |
0.087 |
| 118 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.196 |
-0.037 |
| 119 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.576 |
0.034 |
| 120 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.442 |
0.022 |
| 121 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
-0.426 |
0.052 |
| 122 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.397 |
0.243 |
| 123 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.384 |
0.099 |
| 124 |
263384_at |
AT2G40130
|
SMXL8, SMAX1-like 8 |
-0.377 |
0.007 |
| 125 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.374 |
0.078 |
| 126 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.363 |
-0.289 |
| 127 |
252501_at |
AT3G46880
|
unknown |
-0.358 |
-0.106 |
| 128 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.352 |
0.067 |
| 129 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
-0.341 |
-0.101 |
| 130 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
-0.331 |
0.136 |
| 131 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.321 |
0.067 |
| 132 |
262211_at |
AT1G74930
|
ORA47 |
-0.317 |
0.018 |
| 133 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-0.315 |
0.313 |
| 134 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.302 |
0.204 |
| 135 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.300 |
0.080 |
| 136 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.295 |
0.210 |
| 137 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.290 |
-0.143 |
| 138 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.286 |
-0.403 |
| 139 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.282 |
0.126 |
| 140 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.282 |
-0.001 |
| 141 |
254145_at |
AT4G24700
|
unknown |
-0.282 |
0.052 |
| 142 |
253814_at |
AT4G28290
|
unknown |
-0.281 |
0.012 |
| 143 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.279 |
0.224 |
| 144 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.276 |
0.111 |
| 145 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.276 |
0.149 |
| 146 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.276 |
-0.195 |
| 147 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.275 |
0.880 |
| 148 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.275 |
0.132 |
| 149 |
247753_at |
AT5G59070
|
[UDP-Glycosyltransferase superfamily protein] |
-0.274 |
-0.084 |
| 150 |
247324_at |
AT5G64190
|
unknown |
-0.271 |
-0.261 |
| 151 |
256096_at |
AT1G13650
|
unknown |
-0.271 |
0.029 |
| 152 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.269 |
-0.071 |
| 153 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.269 |
0.072 |
| 154 |
246997_at |
AT5G67390
|
unknown |
-0.269 |
-0.323 |
| 155 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.268 |
-0.180 |
| 156 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.268 |
-0.610 |
| 157 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.268 |
0.082 |
| 158 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.267 |
0.027 |
| 159 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.266 |
0.107 |
| 160 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.266 |
0.044 |
| 161 |
261772_at |
AT1G76240
|
unknown |
-0.262 |
-0.492 |
| 162 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.261 |
-0.059 |
| 163 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.261 |
0.166 |
| 164 |
249932_at |
AT5G22390
|
unknown |
-0.261 |
0.141 |
| 165 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.260 |
0.295 |
| 166 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.259 |
0.180 |
| 167 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.257 |
0.437 |
| 168 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.256 |
0.302 |
| 169 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.254 |
-0.203 |
| 170 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.253 |
0.007 |
| 171 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.253 |
-0.054 |
| 172 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
-0.252 |
0.647 |
| 173 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.252 |
0.001 |
| 174 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.251 |
-0.043 |
| 175 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.251 |
0.233 |
| 176 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.250 |
0.203 |
| 177 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.250 |
-0.061 |
| 178 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.250 |
0.108 |
| 179 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.247 |
0.150 |
| 180 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.246 |
0.130 |
| 181 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
-0.245 |
-0.126 |
| 182 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.243 |
-0.048 |
| 183 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.242 |
-0.045 |
| 184 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.240 |
-0.037 |
| 185 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.240 |
0.081 |
| 186 |
248646_at |
AT5G49100
|
unknown |
-0.237 |
0.075 |
| 187 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.237 |
-0.330 |
| 188 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.237 |
0.215 |
| 189 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.236 |
0.147 |
| 190 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.234 |
-0.102 |
| 191 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.234 |
-0.147 |
| 192 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.231 |
-0.318 |
| 193 |
249726_at |
AT5G35480
|
unknown |
-0.231 |
0.094 |
| 194 |
265732_at |
AT2G01300
|
unknown |
-0.229 |
-0.223 |
| 195 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.228 |
0.079 |
| 196 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.224 |
0.081 |
| 197 |
261319_at |
AT1G53090
|
SPA4, SPA1-related 4 |
-0.223 |
-0.000 |
| 198 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.223 |
-0.262 |
| 199 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.220 |
-0.019 |
| 200 |
260696_at |
AT1G32520
|
unknown |
-0.219 |
0.095 |
| 201 |
259535_at |
AT1G12280
|
SUMM2, suppressor of mkk1 mkk2 2 |
-0.218 |
0.148 |
| 202 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.218 |
0.052 |
| 203 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.217 |
-0.055 |
| 204 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.217 |
0.056 |
| 205 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.216 |
0.058 |
| 206 |
248028_at |
AT5G55620
|
unknown |
-0.216 |
0.291 |
| 207 |
252619_at |
AT3G45210
|
unknown |
-0.216 |
0.191 |
| 208 |
253859_at |
AT4G27657
|
unknown |
-0.214 |
0.660 |
| 209 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.214 |
0.128 |
| 210 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.213 |
0.083 |
| 211 |
252010_at |
AT3G52740
|
unknown |
-0.212 |
0.121 |
| 212 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.210 |
0.019 |
| 213 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
-0.209 |
0.129 |
| 214 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.206 |
0.046 |
| 215 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
-0.205 |
0.245 |
| 216 |
257925_at |
AT3G23170
|
unknown |
-0.205 |
0.279 |
| 217 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
-0.203 |
0.197 |
| 218 |
266814_at |
AT2G44910
|
ATHB-4, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 4, ATHB4, homeobox-leucine zipper protein 4, HB4, homeobox-leucine zipper protein 4 |
-0.202 |
0.467 |
| 219 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.200 |
0.112 |
| 220 |
245084_at |
AT2G23290
|
AtMYB70, myb domain protein 70, MYB70, myb domain protein 70 |
-0.199 |
-0.099 |
| 221 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.198 |
0.799 |
| 222 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.198 |
-0.398 |
| 223 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.198 |
0.154 |
| 224 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
-0.198 |
0.135 |
| 225 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.197 |
0.396 |
| 226 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.197 |
0.113 |
| 227 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.196 |
0.053 |
| 228 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.194 |
0.166 |
| 229 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.192 |
0.039 |
| 230 |
252073_at |
AT3G51750
|
unknown |
-0.189 |
-0.003 |
| 231 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.188 |
0.016 |
| 232 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
-0.187 |
0.109 |
| 233 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.187 |
-0.110 |
| 234 |
247754_at |
AT5G59080
|
unknown |
-0.185 |
-0.107 |
| 235 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.185 |
-0.237 |