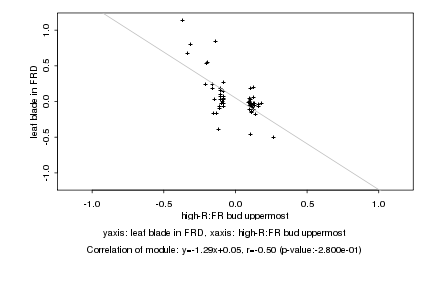
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
high-R:FR bud uppermost |
Log2 signal ratio
leaf blade in FRD |
| 1 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.260 |
-0.491 |
| 2 |
250154_at |
AT5G15140
|
[Galactose mutarotase-like superfamily protein] |
0.177 |
-0.017 |
| 3 |
250783_at |
AT5G05260
|
CYP79A2, cytochrome p450 79a2 |
0.160 |
-0.056 |
| 4 |
263528_at |
AT2G24800
|
[Peroxidase superfamily protein] |
0.157 |
-0.035 |
| 5 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.139 |
-0.171 |
| 6 |
246963_at |
AT5G24820
|
[Eukaryotic aspartyl protease family protein] |
0.130 |
-0.052 |
| 7 |
265984_at |
AT2G24210
|
TPS10, terpene synthase 10 |
0.129 |
-0.014 |
| 8 |
260430_at |
AT1G68200
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.126 |
-0.102 |
| 9 |
259527_at |
AT1G12600
|
[UDP-N-acetylglucosamine (UAA) transporter family] |
0.125 |
0.068 |
| 10 |
250574_at |
AT5G08230
|
[Tudor/PWWP/MBT domain-containing protein] |
0.123 |
-0.024 |
| 11 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.120 |
0.197 |
| 12 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.118 |
-0.079 |
| 13 |
254248_at |
AT4G23270
|
CRK19, cysteine-rich RLK (RECEPTOR-like protein kinase) 19 |
0.111 |
-0.069 |
| 14 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.110 |
-0.147 |
| 15 |
251843_x_at |
AT3G54590
|
ATHRGP1, hydroxyproline-rich glycoprotein, HRGP1, hydroxyproline-rich glycoprotein |
0.110 |
-0.030 |
| 16 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.108 |
-0.127 |
| 17 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.105 |
-0.041 |
| 18 |
250068_at |
AT5G17960
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.105 |
-0.046 |
| 19 |
253641_at |
AT4G29980
|
unknown |
0.104 |
-0.026 |
| 20 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.104 |
-0.457 |
| 21 |
247796_at |
AT5G58782
|
AtcPT6, cPT6, cis-prenyltransferase 6 |
0.104 |
-0.016 |
| 22 |
256334_at |
AT1G72125
|
AtNPF5.13, NPF5.13, NRT1/ PTR family 5.13 |
0.103 |
0.042 |
| 23 |
254408_at |
AT4G21390
|
B120 |
0.102 |
0.184 |
| 24 |
247264_at |
AT5G64530
|
ANAC104, Arabidopsis NAC domain containing protein 104, XND1, xylem NAC domain 1 |
0.096 |
-0.005 |
| 25 |
252253_at |
AT3G49300
|
[proline-rich family protein] |
0.095 |
-0.045 |
| 26 |
245300_at |
AT4G16350
|
CBL6, calcineurin B-like protein 6, SCABP2, SOS3-LIKE CALCIUM BINDING PROTEIN 2 |
0.094 |
-0.108 |
| 27 |
259139_at |
AT3G10240
|
[F-box and associated interaction domains-containing protein] |
0.094 |
-0.048 |
| 28 |
246859_at |
AT5G25950
|
unknown |
0.093 |
0.009 |
| 29 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.092 |
0.049 |
| 30 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.091 |
-0.004 |
| 31 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.371 |
1.149 |
| 32 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.335 |
0.679 |
| 33 |
251012_at |
AT5G02580
|
unknown |
-0.314 |
0.805 |
| 34 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
-0.215 |
0.245 |
| 35 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.203 |
0.533 |
| 36 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.199 |
0.557 |
| 37 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.167 |
0.192 |
| 38 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
-0.162 |
0.248 |
| 39 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.156 |
-0.166 |
| 40 |
254574_at |
AT4G19430
|
unknown |
-0.147 |
0.029 |
| 41 |
265276_at |
AT2G28400
|
unknown |
-0.146 |
0.842 |
| 42 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.133 |
-0.160 |
| 43 |
263829_at |
AT2G40435
|
unknown |
-0.119 |
-0.380 |
| 44 |
246734_at |
AT5G27680
|
RECQSIM, RECQ helicase SIM |
-0.114 |
-0.065 |
| 45 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.112 |
-0.085 |
| 46 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
-0.111 |
0.159 |
| 47 |
248276_at |
AT5G53550
|
ATYSL3, YELLOW STRIPE LIKE 3, YSL3, YELLOW STRIPE like 3 |
-0.111 |
0.110 |
| 48 |
245588_at |
AT4G15030
|
unknown |
-0.109 |
0.076 |
| 49 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.107 |
0.191 |
| 50 |
249763_at |
AT5G24010
|
[Protein kinase superfamily protein] |
-0.106 |
0.036 |
| 51 |
264918_at |
AT1G60530
|
DRP4A, Dynamin related protein 4A |
-0.099 |
-0.023 |
| 52 |
260199_at |
AT1G67590
|
[Remorin family protein] |
-0.094 |
0.010 |
| 53 |
264684_at |
AT1G65590
|
ATHEX1, HEXO3, beta-hexosaminidase 3 |
-0.089 |
0.050 |
| 54 |
252677_at |
AT3G44320
|
AtNIT3, NITRILASE 3, NIT3, nitrilase 3 |
-0.088 |
0.147 |
| 55 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
-0.088 |
-0.061 |
| 56 |
264869_at |
AT1G24350
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
-0.088 |
0.033 |
| 57 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
-0.088 |
0.269 |
| 58 |
265194_at |
AT1G05010
|
ACO4, ethylene forming enzyme, EAT1, EFE, ethylene-forming enzyme |
-0.086 |
0.077 |
| 59 |
266706_at |
AT2G03320
|
unknown |
-0.085 |
-0.022 |