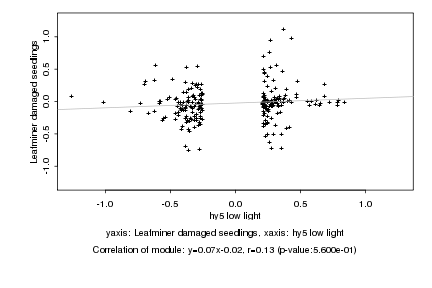
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
hy5 low light |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
250135_at |
AT5G15360
|
unknown |
0.836 |
-0.011 |
| 2 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
0.788 |
0.020 |
| 3 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
0.785 |
-0.013 |
| 4 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
0.782 |
-0.059 |
| 5 |
262421_at |
AT1G50290
|
unknown |
0.719 |
-0.009 |
| 6 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
0.684 |
0.275 |
| 7 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.681 |
0.090 |
| 8 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
0.648 |
-0.020 |
| 9 |
257417_at |
AT1G10110
|
[F-box family protein] |
0.646 |
-0.054 |
| 10 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
0.619 |
0.021 |
| 11 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
0.610 |
-0.043 |
| 12 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.580 |
0.001 |
| 13 |
245245_at |
AT1G44318
|
hemb2 |
0.566 |
-0.052 |
| 14 |
252663_at |
AT3G44070
|
[Glycosyl hydrolase family 35 protein] |
0.551 |
0.002 |
| 15 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
0.471 |
0.310 |
| 16 |
265246_at |
AT2G43050
|
ATPMEPCRD |
0.466 |
0.074 |
| 17 |
265327_at |
AT2G18210
|
unknown |
0.465 |
0.117 |
| 18 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
0.427 |
-0.007 |
| 19 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.424 |
0.989 |
| 20 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
0.415 |
0.027 |
| 21 |
247474_at |
AT5G62280
|
unknown |
0.412 |
-0.400 |
| 22 |
254743_at |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
0.396 |
0.007 |
| 23 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.392 |
0.189 |
| 24 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.388 |
-0.416 |
| 25 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
0.380 |
0.103 |
| 26 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
0.373 |
0.050 |
| 27 |
259478_at |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
0.369 |
-0.001 |
| 28 |
246814_at |
AT5G27200
|
ACP5, acyl carrier protein 5 |
0.369 |
0.033 |
| 29 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.364 |
1.122 |
| 30 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.355 |
0.469 |
| 31 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.353 |
-0.054 |
| 32 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
0.353 |
-0.720 |
| 33 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
0.351 |
-0.498 |
| 34 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.349 |
-0.049 |
| 35 |
262659_at |
AT1G14240
|
[GDA1/CD39 nucleoside phosphatase family protein] |
0.346 |
0.011 |
| 36 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.340 |
-0.156 |
| 37 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.337 |
0.091 |
| 38 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
0.333 |
0.052 |
| 39 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.327 |
-0.076 |
| 40 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
0.327 |
0.034 |
| 41 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
0.326 |
-0.052 |
| 42 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.322 |
-0.025 |
| 43 |
252765_at |
AT3G42800
|
unknown |
0.320 |
-0.176 |
| 44 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
0.319 |
0.060 |
| 45 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.314 |
0.562 |
| 46 |
261878_at |
AT1G50560
|
CYP705A25, cytochrome P450, family 705, subfamily A, polypeptide 25 |
0.309 |
0.039 |
| 47 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
0.308 |
-0.009 |
| 48 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.307 |
0.210 |
| 49 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.301 |
-0.365 |
| 50 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
0.301 |
0.007 |
| 51 |
248509_at |
AT5G50335
|
unknown |
0.296 |
0.063 |
| 52 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.295 |
0.043 |
| 53 |
262665_at |
AT1G14070
|
FUT7, fucosyltransferase 7 |
0.292 |
0.328 |
| 54 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.291 |
-0.508 |
| 55 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
0.286 |
-0.056 |
| 56 |
262855_at |
AT1G20860
|
PHT1;8, phosphate transporter 1;8 |
0.282 |
-0.025 |
| 57 |
267626_at |
AT2G42250
|
CYP712A1, cytochrome P450, family 712, subfamily A, polypeptide 1 |
0.281 |
-0.069 |
| 58 |
246168_at |
AT5G32460
|
[Transcriptional factor B3 family protein] |
0.279 |
-0.016 |
| 59 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
0.278 |
-0.020 |
| 60 |
247476_at |
AT5G62330
|
unknown |
0.277 |
-0.068 |
| 61 |
250327_at |
AT5G12050
|
unknown |
0.275 |
-0.252 |
| 62 |
248394_at |
AT5G52070
|
[Agenet domain-containing protein] |
0.274 |
0.012 |
| 63 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
0.271 |
0.023 |
| 64 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.271 |
-0.712 |
| 65 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
0.270 |
0.157 |
| 66 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.269 |
-0.042 |
| 67 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.268 |
0.951 |
| 68 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
0.266 |
0.540 |
| 69 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
0.260 |
-0.071 |
| 70 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
0.258 |
-0.619 |
| 71 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.257 |
0.762 |
| 72 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.253 |
-0.148 |
| 73 |
252222_at |
AT3G49845
|
WIH3, WINDHOSE 3 |
0.253 |
0.004 |
| 74 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
0.250 |
-0.112 |
| 75 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
0.246 |
-0.327 |
| 76 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.246 |
-0.011 |
| 77 |
251023_at |
AT5G02170
|
[Transmembrane amino acid transporter family protein] |
0.246 |
0.391 |
| 78 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
0.245 |
-0.216 |
| 79 |
259520_at |
AT1G12320
|
unknown |
0.244 |
-0.010 |
| 80 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.243 |
-0.500 |
| 81 |
250699_at |
AT5G06820
|
SRF2, STRUBBELIG-receptor family 2 |
0.242 |
-0.009 |
| 82 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.242 |
-0.137 |
| 83 |
249461_at |
AT5G39640
|
[Putative endonuclease or glycosyl hydrolase] |
0.241 |
0.026 |
| 84 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.241 |
-0.311 |
| 85 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
0.240 |
0.241 |
| 86 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.235 |
-0.103 |
| 87 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
0.235 |
-0.066 |
| 88 |
262233_at |
AT1G48310
|
CHA18, CHR18, chromatin remodeling factor18 |
0.231 |
-0.049 |
| 89 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
0.231 |
-0.289 |
| 90 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
0.230 |
0.040 |
| 91 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.230 |
-0.351 |
| 92 |
259528_at |
AT1G12330
|
unknown |
0.224 |
-0.534 |
| 93 |
249802_at |
AT5G23770
|
unknown |
0.224 |
-0.101 |
| 94 |
256935_at |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.222 |
0.074 |
| 95 |
245554_at |
AT4G15380
|
CYP705A4, cytochrome P450, family 705, subfamily A, polypeptide 4 |
0.220 |
0.002 |
| 96 |
249767_at |
AT5G24090
|
ATCHIA, chitinase A, CHIA, chitinase A |
0.219 |
0.442 |
| 97 |
264347_at |
AT1G12040
|
LRX1, leucine-rich repeat/extensin 1 |
0.219 |
-0.034 |
| 98 |
261772_at |
AT1G76240
|
unknown |
0.218 |
0.097 |
| 99 |
257670_at |
AT3G20340
|
unknown |
0.218 |
0.311 |
| 100 |
249603_at |
AT5G37210
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.218 |
0.025 |
| 101 |
259964_at |
AT1G53680
|
ATGSTU28, glutathione S-transferase TAU 28, GSTU28, glutathione S-transferase TAU 28 |
0.217 |
-0.178 |
| 102 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.217 |
-0.079 |
| 103 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.216 |
0.452 |
| 104 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.214 |
0.496 |
| 105 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.214 |
-0.214 |
| 106 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
0.213 |
-0.381 |
| 107 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
0.213 |
-0.330 |
| 108 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
0.212 |
0.064 |
| 109 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
0.212 |
-0.071 |
| 110 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.211 |
0.082 |
| 111 |
261205_at |
AT1G12790
|
unknown |
0.211 |
0.136 |
| 112 |
263258_at |
AT1G10540
|
ATNAT8, NAT8, nucleobase-ascorbate transporter 8 |
0.210 |
0.015 |
| 113 |
264590_at |
AT2G17710
|
unknown |
0.209 |
-0.084 |
| 114 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.209 |
-0.149 |
| 115 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.209 |
0.697 |
| 116 |
250952_at |
AT5G03570
|
ATIREG2, ARABIDOPSIS THALIANA IRON-REGULATED PROTEIN 2, FPN2, FERROPORTIN 2, IREG2, iron regulated 2 |
0.207 |
-0.023 |
| 117 |
254777_at |
AT4G12960
|
AtGILT, GILT, gamma-interferon-responsive lysosomal thiol reductase |
0.205 |
-0.031 |
| 118 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
0.202 |
-0.007 |
| 119 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
-1.267 |
0.078 |
| 120 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.018 |
-0.009 |
| 121 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.811 |
-0.143 |
| 122 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.732 |
-0.027 |
| 123 |
246888_at |
AT5G26270
|
unknown |
-0.701 |
0.273 |
| 124 |
250931_at |
AT5G03200
|
LUL1, LOG2-LIKE UBIQUITIN LIGASE1 |
-0.697 |
0.322 |
| 125 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.676 |
-0.181 |
| 126 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-0.624 |
-0.145 |
| 127 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
-0.623 |
0.329 |
| 128 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.619 |
0.561 |
| 129 |
259529_at |
AT1G12400
|
[Nucleotide excision repair, TFIIH, subunit TTDA] |
-0.588 |
0.005 |
| 130 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.585 |
-0.017 |
| 131 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.582 |
0.004 |
| 132 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.568 |
-0.287 |
| 133 |
262569_at |
AT1G15180
|
[MATE efflux family protein] |
-0.559 |
-0.257 |
| 134 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.541 |
-0.243 |
| 135 |
264658_at |
AT1G09910
|
[Rhamnogalacturonate lyase family protein] |
-0.529 |
0.043 |
| 136 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.510 |
0.065 |
| 137 |
264958_at |
AT1G76960
|
unknown |
-0.489 |
0.352 |
| 138 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.467 |
-0.170 |
| 139 |
262787_at |
AT1G10730
|
AP1M1, adaptor protein-1 mu-adaptin 1 |
-0.464 |
0.040 |
| 140 |
246079_at |
AT5G20450
|
unknown |
-0.463 |
-0.268 |
| 141 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
-0.455 |
0.048 |
| 142 |
252659_at |
AT3G44430
|
unknown |
-0.447 |
-0.076 |
| 143 |
266015_at |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
-0.445 |
-0.168 |
| 144 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.443 |
-0.211 |
| 145 |
245126_at |
AT2G47460
|
ATMYB12, MYB DOMAIN PROTEIN 12, MYB12, myb domain protein 12, PFG1, PRODUCTION OF FLAVONOL GLYCOSIDES 1 |
-0.439 |
-0.002 |
| 146 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-0.427 |
-0.041 |
| 147 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.427 |
-0.003 |
| 148 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.426 |
-0.107 |
| 149 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.421 |
-0.425 |
| 150 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.421 |
-0.113 |
| 151 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.419 |
-0.129 |
| 152 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.411 |
-0.383 |
| 153 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.409 |
-0.057 |
| 154 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.408 |
-0.116 |
| 155 |
260502_at |
AT1G47270
|
AtTLP6, tubby like protein 6, TLP6, tubby like protein 6 |
-0.404 |
-0.125 |
| 156 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
-0.403 |
0.143 |
| 157 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
-0.400 |
-0.001 |
| 158 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.390 |
0.300 |
| 159 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.389 |
-0.680 |
| 160 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.388 |
-0.138 |
| 161 |
249331_at |
AT5G40950
|
RPL27, ribosomal protein large subunit 27 |
-0.388 |
-0.087 |
| 162 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
-0.388 |
-0.251 |
| 163 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.387 |
-0.100 |
| 164 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.383 |
-0.319 |
| 165 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.383 |
0.529 |
| 166 |
252937_at |
AT4G39180
|
ATSEC14, ARABIDOPSIS THALIANA SECRETION 14, SEC14, SECRETION 14 |
-0.383 |
-0.059 |
| 167 |
246411_at |
AT1G57770
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.381 |
-0.225 |
| 168 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.377 |
0.148 |
| 169 |
256924_at |
AT3G29590
|
AT5MAT |
-0.373 |
0.013 |
| 170 |
260465_at |
AT1G10910
|
EMB3103, EMBRYO DEFECTIVE 3103 |
-0.369 |
-0.291 |
| 171 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.368 |
0.065 |
| 172 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.364 |
-0.420 |
| 173 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
-0.364 |
-0.003 |
| 174 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.364 |
-0.745 |
| 175 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.362 |
0.027 |
| 176 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.362 |
0.197 |
| 177 |
246818_at |
AT5G27270
|
EMB976, EMBRYO DEFECTIVE 976 |
-0.360 |
-0.306 |
| 178 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.360 |
-0.461 |
| 179 |
249612_at |
AT5G37290
|
[ARM repeat superfamily protein] |
-0.349 |
-0.250 |
| 180 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
-0.347 |
-0.075 |
| 181 |
265142_at |
AT1G51360
|
ATDABB1, DIMERIC A/B BARREL DOMAINS-PROTEIN 1, DABB1, dimeric A/B barrel domainS-protein 1 |
-0.342 |
-0.082 |
| 182 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.341 |
0.207 |
| 183 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.340 |
-0.277 |
| 184 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
-0.338 |
0.285 |
| 185 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.336 |
0.086 |
| 186 |
257497_at |
AT1G51430
|
unknown |
-0.334 |
-0.003 |
| 187 |
246087_at |
AT5G20580
|
unknown |
-0.333 |
-0.106 |
| 188 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
-0.332 |
-0.164 |
| 189 |
250740_at |
AT5G05760
|
ATSED5, ATSYP31, SED5, T-SNARE SED 5, SYP31, syntaxin of plants 31 |
-0.327 |
0.107 |
| 190 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.322 |
-0.279 |
| 191 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
-0.316 |
-0.396 |
| 192 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.316 |
0.073 |
| 193 |
261602_at |
AT1G49630
|
ATPREP2, presequence protease 2, PREP2, presequence protease 2 |
-0.316 |
-0.218 |
| 194 |
252444_at |
AT3G46980
|
PHT4;3, phosphate transporter 4;3 |
-0.316 |
-0.068 |
| 195 |
259489_at |
AT1G15790
|
unknown |
-0.315 |
0.249 |
| 196 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.310 |
0.039 |
| 197 |
260966_at |
AT1G12220
|
RPS5, RESISTANT TO P. SYRINGAE 5 |
-0.310 |
-0.040 |
| 198 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.304 |
-0.188 |
| 199 |
249811_at |
AT5G23760
|
[Copper transport protein family] |
-0.303 |
-0.284 |
| 200 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.300 |
-0.258 |
| 201 |
245702_at |
AT5G04220
|
ATSYTC, NTMC2T1.3, NTMC2TYPE1.3, SYT3, synaptotagmin 3, SYTC |
-0.300 |
0.272 |
| 202 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.299 |
0.224 |
| 203 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.297 |
0.549 |
| 204 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
-0.291 |
0.265 |
| 205 |
249724_at |
AT5G35450
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.286 |
-0.362 |
| 206 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.285 |
-0.117 |
| 207 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.285 |
-0.004 |
| 208 |
261879_at |
AT1G50520
|
CYP705A27, cytochrome P450, family 705, subfamily A, polypeptide 27 |
-0.285 |
-0.022 |
| 209 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.282 |
-0.319 |
| 210 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.280 |
-0.730 |
| 211 |
264824_at |
AT1G03420
|
Sadhu4-2, sadhu non-coding retrotransposon 4-2 |
-0.279 |
-0.063 |
| 212 |
262485_at |
AT1G21730
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.278 |
-0.097 |
| 213 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.278 |
0.099 |
| 214 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
-0.275 |
0.200 |
| 215 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
-0.271 |
-0.261 |
| 216 |
245859_at |
AT5G28290
|
ATNEK3, NIMA-related kinase 3, NEK3, NIMA-related kinase 3 |
-0.271 |
0.035 |
| 217 |
256137_at |
AT1G48690
|
[Auxin-responsive GH3 family protein] |
-0.271 |
-0.056 |
| 218 |
246536_at |
AT5G15920
|
EMB2782, EMBRYO DEFECTIVE 2782, SMC5, structural maintenance of chromosomes 5 |
-0.270 |
-0.230 |
| 219 |
261447_at |
AT1G21160
|
[eukaryotic translation initiation factor 2 (eIF-2) family protein] |
-0.270 |
-0.081 |
| 220 |
260660_at |
AT1G19485
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.269 |
-0.168 |
| 221 |
260685_at |
AT1G17650
|
AtGLYR2, GLYR2, glyoxylate reductase 2, GR2, GLYOXYLATE REDUCTASE 2 |
-0.269 |
-0.351 |
| 222 |
252593_at |
AT3G45590
|
ATSEN1, splicing endonuclease 1, SEN1, splicing endonuclease 1 |
-0.268 |
0.107 |
| 223 |
252700_at |
AT3G43690
|
[copia-like retrotransposon family protein, has a 1.4e-29 P-value blast match to gb|AAG52950.1| putative envelope protein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
-0.268 |
-0.096 |
| 224 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.267 |
0.017 |
| 225 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
-0.266 |
-0.038 |
| 226 |
249622_at |
AT5G37550
|
unknown |
-0.266 |
-0.044 |
| 227 |
254437_s_at |
AT3G06433
|
[pseudogene] |
-0.265 |
-0.097 |
| 228 |
265441_at |
AT2G20870
|
[cell wall protein precursor, putative] |
-0.265 |
0.274 |
| 229 |
245079_at |
AT2G23330
|
[copia-like retrotransposon family, has a 3.9e-195 P-value blast match to GB:AAA57005 Hopscotch polyprotein (Ty1_Copia-element) (Zea mays)] |
-0.265 |
0.124 |
| 230 |
259411_at |
AT1G13410
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.262 |
0.043 |
| 231 |
252572_at |
AT3G45290
|
ATMLO3, MILDEW RESISTANCE LOCUS O 3, MLO3, MILDEW RESISTANCE LOCUS O 3 |
-0.261 |
0.121 |
| 232 |
249186_at |
AT5G43040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.260 |
-0.037 |
| 233 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.256 |
-0.128 |
| 234 |
249355_at |
AT5G40500
|
unknown |
-0.255 |
0.137 |
| 235 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.254 |
-0.275 |