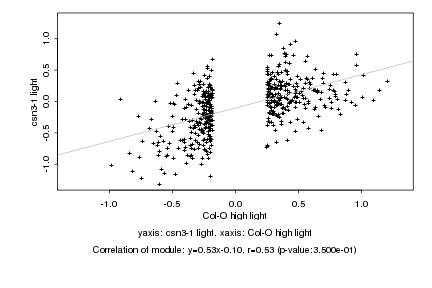
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-O high light |
Log2 signal ratio
csn3-1 light |
| 1 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
1.205 |
0.328 |
| 2 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
1.140 |
0.178 |
| 3 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.093 |
0.018 |
| 4 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
1.014 |
0.419 |
| 5 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
1.008 |
0.080 |
| 6 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.954 |
0.590 |
| 7 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.953 |
0.759 |
| 8 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.948 |
-0.049 |
| 9 |
248218_at |
AT5G53710
|
unknown |
0.915 |
-0.004 |
| 10 |
249771_at |
AT5G24080
|
[Protein kinase superfamily protein] |
0.892 |
0.177 |
| 11 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.875 |
0.123 |
| 12 |
253799_at |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
0.873 |
0.026 |
| 13 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.873 |
0.316 |
| 14 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.827 |
-0.198 |
| 15 |
265024_at |
AT1G24600
|
unknown |
0.811 |
-0.124 |
| 16 |
250826_at |
AT5G05220
|
unknown |
0.808 |
0.034 |
| 17 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.797 |
0.432 |
| 18 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.791 |
0.119 |
| 19 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.788 |
0.083 |
| 20 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.779 |
0.169 |
| 21 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.774 |
0.431 |
| 22 |
264580_at |
AT1G05340
|
unknown |
0.772 |
0.190 |
| 23 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.762 |
-0.098 |
| 24 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.756 |
-0.009 |
| 25 |
256789_at |
AT3G13672
|
SINA2 |
0.747 |
0.259 |
| 26 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.745 |
0.203 |
| 27 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.744 |
0.060 |
| 28 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.710 |
-0.092 |
| 29 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.704 |
-0.060 |
| 30 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.694 |
0.302 |
| 31 |
263881_at |
AT2G21820
|
unknown |
0.692 |
0.450 |
| 32 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.690 |
0.368 |
| 33 |
252882_at |
AT4G39675
|
unknown |
0.682 |
-0.458 |
| 34 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.680 |
0.148 |
| 35 |
251156_at |
AT3G63060
|
EDL3, EID1-like 3 |
0.675 |
0.038 |
| 36 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.657 |
-0.223 |
| 37 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.651 |
0.156 |
| 38 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.649 |
0.168 |
| 39 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.648 |
0.189 |
| 40 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.634 |
0.524 |
| 41 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.632 |
0.178 |
| 42 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.628 |
-0.094 |
| 43 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.621 |
-0.118 |
| 44 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.621 |
0.187 |
| 45 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.615 |
0.187 |
| 46 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.605 |
0.370 |
| 47 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.595 |
0.287 |
| 48 |
248205_at |
AT5G54300
|
unknown |
0.587 |
0.222 |
| 49 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.585 |
0.306 |
| 50 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.579 |
-0.031 |
| 51 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.576 |
-0.430 |
| 52 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.575 |
0.004 |
| 53 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.567 |
0.354 |
| 54 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.566 |
0.725 |
| 55 |
257186_at |
AT3G13130
|
unknown |
0.558 |
0.370 |
| 56 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.555 |
0.300 |
| 57 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.553 |
0.644 |
| 58 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.552 |
0.054 |
| 59 |
255943_at |
AT1G22370
|
AtUGT85A5, UDP-glucosyl transferase 85A5, UGT85A5, UDP-glucosyl transferase 85A5 |
0.549 |
-0.103 |
| 60 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.544 |
-0.032 |
| 61 |
249614_at |
AT5G37300
|
WSD1 |
0.539 |
0.058 |
| 62 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.538 |
0.212 |
| 63 |
260668_at |
AT1G19530
|
unknown |
0.536 |
-0.321 |
| 64 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.525 |
0.099 |
| 65 |
260420_at |
AT1G69610
|
[FUNCTIONS IN: structural constituent of ribosome] |
0.519 |
0.165 |
| 66 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.513 |
0.358 |
| 67 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.508 |
0.128 |
| 68 |
265828_at |
AT2G14520
|
unknown |
0.505 |
0.000 |
| 69 |
261088_at |
AT1G07590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.487 |
0.075 |
| 70 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.485 |
0.410 |
| 71 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.485 |
0.244 |
| 72 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.485 |
-0.280 |
| 73 |
250158_at |
AT5G15190
|
unknown |
0.484 |
-0.007 |
| 74 |
260005_at |
AT1G67920
|
unknown |
0.484 |
0.168 |
| 75 |
263492_at |
AT2G42560
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.483 |
0.070 |
| 76 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
0.481 |
0.291 |
| 77 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.476 |
0.971 |
| 78 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.475 |
-0.090 |
| 79 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.475 |
0.131 |
| 80 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
0.475 |
0.089 |
| 81 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.474 |
0.199 |
| 82 |
264774_at |
AT1G22890
|
unknown |
0.472 |
-0.466 |
| 83 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.471 |
-0.009 |
| 84 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.465 |
0.072 |
| 85 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.459 |
0.749 |
| 86 |
255527_at |
AT4G02360
|
unknown |
0.454 |
0.245 |
| 87 |
267206_at |
AT2G30830
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.448 |
0.032 |
| 88 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
0.443 |
0.004 |
| 89 |
249454_at |
AT5G39520
|
unknown |
0.436 |
0.921 |
| 90 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.435 |
0.361 |
| 91 |
256603_at |
AT3G28270
|
unknown |
0.432 |
0.007 |
| 92 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.431 |
-0.326 |
| 93 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
0.429 |
0.284 |
| 94 |
248505_at |
AT5G50360
|
unknown |
0.428 |
0.145 |
| 95 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.425 |
0.612 |
| 96 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.424 |
0.612 |
| 97 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.423 |
-0.015 |
| 98 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.423 |
-0.025 |
| 99 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
0.422 |
-0.025 |
| 100 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.421 |
0.280 |
| 101 |
255602_at |
AT4G01026
|
PYL7, PYR1-like 7, RCAR2, regulatory components of ABA receptor 2 |
0.417 |
-0.139 |
| 102 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.413 |
0.480 |
| 103 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.411 |
-0.619 |
| 104 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
0.410 |
0.117 |
| 105 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.408 |
0.179 |
| 106 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
0.408 |
0.438 |
| 107 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.407 |
-0.077 |
| 108 |
263265_at |
AT2G38820
|
unknown |
0.407 |
0.045 |
| 109 |
248959_at |
AT5G45630
|
unknown |
0.402 |
0.051 |
| 110 |
264958_at |
AT1G76960
|
unknown |
0.401 |
0.763 |
| 111 |
260522_x_at |
AT2G41730
|
unknown |
0.399 |
0.486 |
| 112 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.399 |
0.295 |
| 113 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.396 |
0.241 |
| 114 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.395 |
-0.123 |
| 115 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.395 |
0.761 |
| 116 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.393 |
0.507 |
| 117 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.393 |
0.722 |
| 118 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.393 |
0.190 |
| 119 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.390 |
0.637 |
| 120 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.388 |
0.265 |
| 121 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.385 |
0.065 |
| 122 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.384 |
0.778 |
| 123 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.380 |
-0.089 |
| 124 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.376 |
-0.175 |
| 125 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.376 |
0.851 |
| 126 |
258275_at |
AT3G15760
|
unknown |
0.372 |
-0.106 |
| 127 |
265059_at |
AT1G52080
|
AR791 |
0.367 |
0.320 |
| 128 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.366 |
-0.186 |
| 129 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
0.362 |
-0.036 |
| 130 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.362 |
0.469 |
| 131 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.361 |
0.246 |
| 132 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.361 |
-0.364 |
| 133 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
0.361 |
-0.313 |
| 134 |
255348_at |
AT4G03820
|
unknown |
0.359 |
-0.032 |
| 135 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.359 |
0.270 |
| 136 |
265276_at |
AT2G28400
|
unknown |
0.359 |
0.066 |
| 137 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.355 |
0.057 |
| 138 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
0.355 |
0.264 |
| 139 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.353 |
0.108 |
| 140 |
262607_at |
AT1G13990
|
unknown |
0.353 |
0.195 |
| 141 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
0.352 |
0.307 |
| 142 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.352 |
-0.037 |
| 143 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.350 |
0.211 |
| 144 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.349 |
0.021 |
| 145 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.349 |
0.598 |
| 146 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.347 |
1.252 |
| 147 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.344 |
-0.241 |
| 148 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.343 |
0.332 |
| 149 |
248428_at |
AT5G51760
|
AHG1, ABA-hypersensitive germination 1 |
0.340 |
0.262 |
| 150 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.340 |
-0.234 |
| 151 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
0.339 |
0.132 |
| 152 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
0.338 |
0.437 |
| 153 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.336 |
0.052 |
| 154 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.336 |
-0.020 |
| 155 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.335 |
0.559 |
| 156 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.334 |
0.570 |
| 157 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.334 |
-0.193 |
| 158 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
0.332 |
0.464 |
| 159 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.332 |
0.079 |
| 160 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.330 |
0.210 |
| 161 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
0.329 |
-0.233 |
| 162 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.328 |
-0.008 |
| 163 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.326 |
-0.061 |
| 164 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.325 |
0.355 |
| 165 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.325 |
-0.642 |
| 166 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.324 |
-0.041 |
| 167 |
265452_at |
AT2G46510
|
AIB, ABA-inducible BHLH-type transcription factor, ATAIB, ABA-inducible BHLH-type transcription factor, JAM1, JA-associated MYC2-like 1 |
0.324 |
-0.057 |
| 168 |
258262_at |
AT3G15770
|
unknown |
0.321 |
-0.220 |
| 169 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.320 |
1.069 |
| 170 |
260039_at |
AT1G68795
|
CLE12, CLAVATA3/ESR-RELATED 12 |
0.320 |
0.208 |
| 171 |
255872_at |
AT2G30360
|
CIPK11, CBL-INTERACTING PROTEIN KINASE 11, PKS5, PROTEIN KINASE SOS2-LIKE 5, SIP4, SOS3-interacting protein 4, SNRK3.22, SNF1-RELATED PROTEIN KINASE 3.22 |
0.317 |
0.237 |
| 172 |
265481_at |
AT2G15960
|
unknown |
0.315 |
-0.457 |
| 173 |
255787_at |
AT2G33590
|
AtCRL1, CRL1, CCR(Cinnamoyl coA:NADP oxidoreductase)-like 1 |
0.314 |
0.118 |
| 174 |
258789_at |
AT3G04605
|
MUG1, MUSTANG 1 |
0.308 |
-0.185 |
| 175 |
264255_at |
AT1G09140
|
At-SR30, Serine/Arginine-Rich Protein Splicing Factor 30, ATSRP30, SERINE-ARGININE PROTEIN 30, ATSRP30.1, ATSRP30.2, SR30, Serine/Arginine-Rich Protein Splicing Factor 30 |
0.307 |
0.004 |
| 176 |
252872_at |
AT4G40010
|
SNRK2-7, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-7, SNRK2.7, SNF1-related protein kinase 2.7, SRK2F |
0.307 |
-0.012 |
| 177 |
250483_at |
AT5G10300
|
AtHNL, ATMES5, ARABIDOPSIS THALIANA METHYL ESTERASE 5, HNL, HYDROXYNITRILE LYASE, MES5, methyl esterase 5 |
0.306 |
0.203 |
| 178 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.306 |
0.287 |
| 179 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.306 |
0.403 |
| 180 |
252097_at |
AT3G51090
|
unknown |
0.305 |
-0.081 |
| 181 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.305 |
0.009 |
| 182 |
258610_at |
AT3G02875
|
ILR1, IAA-LEUCINE RESISTANT 1 |
0.303 |
0.026 |
| 183 |
251267_at |
AT3G62330
|
[Zinc knuckle (CCHC-type) family protein] |
0.302 |
0.091 |
| 184 |
264661_at |
AT1G09950
|
RAS1, RESPONSE TO ABA AND SALT 1 |
0.301 |
0.094 |
| 185 |
263545_at |
AT2G21560
|
unknown |
0.299 |
-0.151 |
| 186 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
0.298 |
-0.201 |
| 187 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
0.295 |
-0.380 |
| 188 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.294 |
0.126 |
| 189 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.294 |
0.065 |
| 190 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
0.294 |
-0.200 |
| 191 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.293 |
0.476 |
| 192 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
0.290 |
0.080 |
| 193 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
0.290 |
-0.176 |
| 194 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.289 |
0.224 |
| 195 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
0.286 |
-0.330 |
| 196 |
263157_at |
AT1G54100
|
ALDH7B4, aldehyde dehydrogenase 7B4 |
0.285 |
0.022 |
| 197 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.284 |
0.357 |
| 198 |
256433_at |
AT3G10985
|
ATWI-12, ARABIDOPSIS THALIANA WOUND-INDUCED PROTEIN 12, SAG20, senescence associated gene 20, WI12, WOUND-INDUCED PROTEIN 12 |
0.284 |
-0.324 |
| 199 |
261545_at |
AT1G63530
|
unknown |
0.283 |
0.742 |
| 200 |
248248_at |
AT5G53120
|
ATSPDS3, SPERMIDINE SYNTHASE 3, SPDS3, spermidine synthase 3, SPMS |
0.283 |
0.353 |
| 201 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.282 |
-0.008 |
| 202 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.282 |
0.218 |
| 203 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.282 |
0.032 |
| 204 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.281 |
-0.131 |
| 205 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.281 |
0.226 |
| 206 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.280 |
0.380 |
| 207 |
252321_at |
AT3G48510
|
unknown |
0.279 |
-0.094 |
| 208 |
248487_at |
AT5G51070
|
CLPD, ERD1, EARLY RESPONSIVE TO DEHYDRATION 1, SAG15, SENESCENCE ASSOCIATED GENE 15 |
0.279 |
0.062 |
| 209 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.276 |
0.193 |
| 210 |
253800_at |
AT4G28160
|
[hydroxyproline-rich glycoprotein family protein] |
0.275 |
-0.186 |
| 211 |
267483_at |
AT2G02810
|
ATUTR1, UDP-GALACTOSE TRANSPORTER 1, UTR1, UDP-galactose transporter 1 |
0.275 |
0.139 |
| 212 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.274 |
0.356 |
| 213 |
254269_at |
AT4G23050
|
[PAS domain-containing protein tyrosine kinase family protein] |
0.274 |
0.244 |
| 214 |
267429_at |
AT2G34850
|
MEE25, maternal effect embryo arrest 25 |
0.274 |
-0.074 |
| 215 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
0.273 |
-0.090 |
| 216 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.271 |
0.440 |
| 217 |
261399_at |
AT1G79620
|
[Leucine-rich repeat protein kinase family protein] |
0.270 |
-0.334 |
| 218 |
256518_at |
AT1G66080
|
unknown |
0.267 |
-0.185 |
| 219 |
257672_at |
AT3G20300
|
unknown |
0.267 |
-0.166 |
| 220 |
255502_at |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
0.266 |
0.302 |
| 221 |
246781_at |
AT5G27350
|
SFP1 |
0.266 |
-0.019 |
| 222 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
0.265 |
0.325 |
| 223 |
263515_at |
AT2G21640
|
unknown |
0.264 |
0.734 |
| 224 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.263 |
0.153 |
| 225 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
0.263 |
0.125 |
| 226 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.262 |
0.445 |
| 227 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.262 |
0.201 |
| 228 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.262 |
0.210 |
| 229 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.261 |
0.269 |
| 230 |
246779_at |
AT5G27520
|
AtPNC2, PNC2, peroxisomal adenine nucleotide carrier 2 |
0.258 |
0.160 |
| 231 |
260983_at |
AT1G53560
|
[Ribosomal protein L18ae family] |
0.257 |
-0.311 |
| 232 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.257 |
-0.270 |
| 233 |
265411_at |
AT2G16630
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.256 |
0.089 |
| 234 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
0.255 |
0.103 |
| 235 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.253 |
0.492 |
| 236 |
260841_at |
AT1G29195
|
unknown |
0.253 |
0.077 |
| 237 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.253 |
0.120 |
| 238 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
0.253 |
-0.703 |
| 239 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.251 |
0.434 |
| 240 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
0.251 |
-0.699 |
| 241 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.250 |
-0.001 |
| 242 |
258264_at |
AT3G15790
|
ATMBD11, MBD11, methyl-CPG-binding domain 11 |
0.250 |
-0.050 |
| 243 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.249 |
0.517 |
| 244 |
252173_at |
AT3G50650
|
[GRAS family transcription factor] |
0.248 |
-0.380 |
| 245 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
0.247 |
0.549 |
| 246 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.247 |
0.286 |
| 247 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
0.247 |
0.304 |
| 248 |
251858_at |
AT3G54820
|
PIP2;5, plasma membrane intrinsic protein 2;5, PIP2D, PLASMA MEMBRANE INTRINSIC PROTEIN 2D |
0.247 |
0.030 |
| 249 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.246 |
-0.722 |
| 250 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.988 |
-1.007 |
| 251 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.916 |
0.046 |
| 252 |
248509_at |
AT5G50335
|
unknown |
-0.845 |
-0.825 |
| 253 |
247878_at |
AT5G57760
|
unknown |
-0.823 |
-1.103 |
| 254 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.770 |
-0.232 |
| 255 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.762 |
-0.879 |
| 256 |
247474_at |
AT5G62280
|
unknown |
-0.752 |
-1.216 |
| 257 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.740 |
-0.630 |
| 258 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.683 |
-0.426 |
| 259 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.667 |
-0.277 |
| 260 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.654 |
-0.661 |
| 261 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
-0.637 |
0.013 |
| 262 |
250327_at |
AT5G12050
|
unknown |
-0.628 |
-0.475 |
| 263 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.623 |
-0.700 |
| 264 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.618 |
-0.643 |
| 265 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.614 |
-0.853 |
| 266 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.610 |
-1.307 |
| 267 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.607 |
-0.783 |
| 268 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.599 |
-0.545 |
| 269 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.593 |
-1.081 |
| 270 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.574 |
-0.636 |
| 271 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.566 |
-1.133 |
| 272 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.551 |
-0.868 |
| 273 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.544 |
-0.739 |
| 274 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.540 |
-0.848 |
| 275 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
-0.536 |
-0.389 |
| 276 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.526 |
-0.654 |
| 277 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.520 |
-0.022 |
| 278 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.504 |
-0.470 |
| 279 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.500 |
-0.226 |
| 280 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.495 |
-0.024 |
| 281 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
-0.494 |
-0.406 |
| 282 |
245076_at |
AT2G23170
|
GH3.3 |
-0.492 |
-0.895 |
| 283 |
260742_at |
AT1G15050
|
IAA34, indole-3-acetic acid inducible 34 |
-0.484 |
-0.039 |
| 284 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.477 |
-1.153 |
| 285 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.468 |
0.110 |
| 286 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.466 |
0.290 |
| 287 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.456 |
-0.736 |
| 288 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.444 |
-0.608 |
| 289 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.426 |
-0.610 |
| 290 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
-0.422 |
-0.273 |
| 291 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
-0.412 |
-0.096 |
| 292 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.412 |
-0.644 |
| 293 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.410 |
-0.765 |
| 294 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
-0.409 |
-0.598 |
| 295 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.403 |
-0.282 |
| 296 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.400 |
0.048 |
| 297 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.398 |
-0.718 |
| 298 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
-0.395 |
-0.092 |
| 299 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.393 |
-0.984 |
| 300 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.384 |
-0.846 |
| 301 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
-0.377 |
0.190 |
| 302 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.374 |
-0.036 |
| 303 |
263971_at |
AT2G42800
|
AtRLP29, receptor like protein 29, RLP29, receptor like protein 29 |
-0.368 |
-0.309 |
| 304 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.363 |
-0.401 |
| 305 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.362 |
-0.696 |
| 306 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.360 |
-0.862 |
| 307 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.359 |
-0.583 |
| 308 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.355 |
-0.738 |
| 309 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.352 |
-0.060 |
| 310 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.351 |
-0.848 |
| 311 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
-0.350 |
-0.481 |
| 312 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.346 |
-0.903 |
| 313 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.345 |
-0.301 |
| 314 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.343 |
-0.684 |
| 315 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
-0.340 |
-0.419 |
| 316 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
-0.334 |
0.453 |
| 317 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.332 |
-0.756 |
| 318 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.328 |
-0.374 |
| 319 |
261318_at |
AT1G53035
|
unknown |
-0.326 |
0.123 |
| 320 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
-0.326 |
0.262 |
| 321 |
251476_at |
AT3G59670
|
unknown |
-0.323 |
0.180 |
| 322 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
-0.321 |
-0.414 |
| 323 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
-0.319 |
-0.501 |
| 324 |
252765_at |
AT3G42800
|
unknown |
-0.318 |
-0.606 |
| 325 |
251261_at |
AT3G62110
|
[Pectin lyase-like superfamily protein] |
-0.315 |
-0.314 |
| 326 |
263919_at |
AT2G36470
|
unknown |
-0.313 |
0.031 |
| 327 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.310 |
-0.124 |
| 328 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.309 |
-0.349 |
| 329 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.304 |
0.188 |
| 330 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.302 |
-0.641 |
| 331 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
-0.302 |
0.189 |
| 332 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.301 |
-0.536 |
| 333 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.300 |
-0.183 |
| 334 |
262452_at |
AT1G11210
|
unknown |
-0.300 |
0.327 |
| 335 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
-0.299 |
0.210 |
| 336 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
-0.298 |
-0.736 |
| 337 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.296 |
-0.367 |
| 338 |
252204_at |
AT3G50340
|
unknown |
-0.295 |
-0.379 |
| 339 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.290 |
-0.025 |
| 340 |
256097_at |
AT1G13670
|
unknown |
-0.289 |
-0.331 |
| 341 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
-0.288 |
0.280 |
| 342 |
256077_at |
AT1G18090
|
[5'-3' exonuclease family protein] |
-0.287 |
0.013 |
| 343 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.287 |
-0.674 |
| 344 |
258367_at |
AT3G14370
|
WAG2 |
-0.286 |
-0.431 |
| 345 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
-0.286 |
0.273 |
| 346 |
263955_at |
AT2G36010
|
ATE2FA, E2F3, E2F transcription factor 3 |
-0.285 |
0.234 |
| 347 |
251223_at |
AT3G62610
|
ATMYB11, myb domain protein 11, MYB11, myb domain protein 11, PFG2, PRODUCTION OF FLAVONOL GLYCOSIDES 2 |
-0.284 |
0.330 |
| 348 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.284 |
-0.006 |
| 349 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.281 |
-0.350 |
| 350 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.280 |
-0.256 |
| 351 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.277 |
-0.998 |
| 352 |
246487_at |
AT5G16030
|
unknown |
-0.275 |
-0.284 |
| 353 |
250366_at |
AT5G11420
|
unknown |
-0.273 |
-0.440 |
| 354 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
-0.268 |
0.092 |
| 355 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.267 |
-0.484 |
| 356 |
249887_at |
AT5G22310
|
unknown |
-0.266 |
-0.073 |
| 357 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.266 |
-0.191 |
| 358 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
-0.265 |
-0.477 |
| 359 |
246860_at |
AT5G25840
|
unknown |
-0.264 |
-0.902 |
| 360 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.264 |
-0.460 |
| 361 |
248423_at |
AT5G51670
|
unknown |
-0.263 |
-0.451 |
| 362 |
248679_at |
AT5G48830
|
unknown |
-0.262 |
-0.007 |
| 363 |
254073_at |
AT4G25500
|
At-RS40, arginine/serine-rich splicing factor 40, AT-SRP40, ATRSP35, ARABIDOPSIS THALIANA ARGININE/SERINE-RICH SPLICING FACTOR 35, ATRSP40, RS40, arginine/serine-rich splicing factor 40, RSP35, arginine/serine-rich splicing factor 35 |
-0.261 |
-0.012 |
| 364 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
-0.260 |
0.275 |
| 365 |
245743_at |
AT1G51080
|
unknown |
-0.259 |
-0.238 |
| 366 |
261048_at |
AT1G01420
|
UGT72B3, UDP-glucosyl transferase 72B3 |
-0.259 |
-0.185 |
| 367 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
-0.259 |
-0.115 |
| 368 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
-0.257 |
-0.611 |
| 369 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
-0.257 |
0.120 |
| 370 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
-0.257 |
-0.690 |
| 371 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.256 |
-0.501 |
| 372 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.255 |
0.161 |
| 373 |
247319_at |
AT5G64050
|
ATERS, ERS, glutamate tRNA synthetase, OVA3, OVULE ABORTION 3 |
-0.253 |
0.143 |
| 374 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.253 |
-0.751 |
| 375 |
253880_at |
AT4G27590
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.253 |
-0.055 |
| 376 |
252133_at |
AT3G50900
|
unknown |
-0.253 |
-0.169 |
| 377 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.251 |
-0.596 |
| 378 |
253810_at |
AT4G28220
|
NDB1, NAD(P)H dehydrogenase B1 |
-0.249 |
-0.127 |
| 379 |
266760_at |
AT2G46870
|
NGA1, NGATHA1 |
-0.249 |
0.111 |
| 380 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.248 |
0.419 |
| 381 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.246 |
-0.354 |
| 382 |
263931_at |
AT2G36220
|
unknown |
-0.244 |
-0.208 |
| 383 |
247085_at |
AT5G66310
|
[ATP binding microtubule motor family protein] |
-0.244 |
-0.405 |
| 384 |
246419_at |
AT5G17030
|
UGT78D3, UDP-glucosyl transferase 78D3 |
-0.242 |
0.149 |
| 385 |
251141_at |
AT5G01075
|
[Glycosyl hydrolase family 35 protein] |
-0.242 |
-0.772 |
| 386 |
246315_at |
AT3G56870
|
unknown |
-0.241 |
0.035 |
| 387 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
-0.241 |
-0.263 |
| 388 |
258545_at |
AT3G07050
|
NSN1, nucleostemin-like 1 |
-0.240 |
0.134 |
| 389 |
245166_at |
AT2G33170
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
-0.240 |
0.065 |
| 390 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.238 |
-0.350 |
| 391 |
246525_at |
AT5G15840
|
BBX1, B-box domain protein 1, CO, CONSTANS, FG |
-0.238 |
-0.426 |
| 392 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
-0.237 |
-0.018 |
| 393 |
246346_at |
AT3G56810
|
unknown |
-0.235 |
-0.648 |
| 394 |
251244_at |
AT3G62060
|
[Pectinacetylesterase family protein] |
-0.235 |
-0.139 |
| 395 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.235 |
-0.206 |
| 396 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.235 |
-0.405 |
| 397 |
247743_at |
AT5G59010
|
BSK5, brassinosteroid-signaling kinase 5 |
-0.234 |
-0.303 |
| 398 |
248801_at |
AT5G47370
|
HAT2 |
-0.233 |
-0.794 |
| 399 |
257237_at |
AT3G14890
|
[phosphoesterase] |
-0.233 |
-0.075 |
| 400 |
248753_at |
AT5G47630
|
mtACP3, mitochondrial acyl carrier protein 3 |
-0.232 |
0.163 |
| 401 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.231 |
-0.248 |
| 402 |
258204_at |
AT3G13960
|
AtGRF5, growth-regulating factor 5, GRF5, growth-regulating factor 5 |
-0.231 |
-0.336 |
| 403 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.229 |
0.013 |
| 404 |
248940_at |
AT5G45400
|
ATRPA70C, RPA70C |
-0.228 |
0.570 |
| 405 |
262201_at |
AT2G01120
|
ATORC4, ORIGIN RECOGNITION COMPLEX SUBUNIT 4, ORC4, origin recognition complex subunit 4 |
-0.227 |
0.373 |
| 406 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.226 |
-0.269 |
| 407 |
255854_at |
AT1G67050
|
unknown |
-0.226 |
-0.319 |
| 408 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.226 |
-0.600 |
| 409 |
246885_at |
AT5G26230
|
MAKR1, membrane-associated kinase regulator 1 |
-0.226 |
-0.323 |
| 410 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.225 |
0.091 |
| 411 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.225 |
-0.005 |
| 412 |
261516_at |
AT1G71750
|
HGPT, Hypoxanthine-guanine phosphoribosyltransferase |
-0.225 |
-0.229 |
| 413 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
-0.224 |
-0.834 |
| 414 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.223 |
-0.020 |
| 415 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-0.223 |
-0.211 |
| 416 |
263681_at |
AT1G26840
|
ATORC6, ARABIDOPSIS THALIANA ORIGIN RECOGNITION COMPLEX PROTEIN 6, ORC6, origin recognition complex protein 6 |
-0.223 |
0.537 |
| 417 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.221 |
-0.399 |
| 418 |
264521_at |
AT1G10020
|
unknown |
-0.221 |
-0.224 |
| 419 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
-0.221 |
-0.000 |
| 420 |
265931_at |
AT2G18520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.221 |
0.071 |
| 421 |
262149_at |
AT1G52530
|
unknown |
-0.220 |
0.137 |
| 422 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
-0.220 |
0.009 |
| 423 |
248784_at |
AT5G47380
|
unknown |
-0.218 |
-0.519 |
| 424 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
-0.218 |
-0.903 |
| 425 |
252659_at |
AT3G44430
|
unknown |
-0.218 |
0.031 |
| 426 |
265831_at |
AT2G14460
|
unknown |
-0.217 |
0.229 |
| 427 |
258024_at |
AT3G19360
|
[Zinc finger (CCCH-type) family protein] |
-0.217 |
0.165 |
| 428 |
252183_at |
AT3G50740
|
UGT72E1, UDP-glucosyl transferase 72E1 |
-0.217 |
-0.142 |
| 429 |
259845_at |
AT1G73590
|
ATPIN1, ARABIDOPSIS THALIANA PIN-FORMED 1, PIN1, PIN-FORMED 1 |
-0.217 |
-0.341 |
| 430 |
262850_at |
AT1G14920
|
GAI, GIBBERELLIC ACID INSENSITIVE, RGA2, RESTORATION ON GROWTH ON AMMONIA 2 |
-0.216 |
-0.467 |
| 431 |
261058_at |
AT1G01370
|
CENH3, CENTROMERIC HISTONE H3, HTR12 |
-0.216 |
0.228 |
| 432 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
-0.213 |
-0.596 |
| 433 |
259209_at |
AT3G09160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.213 |
0.031 |
| 434 |
255028_at |
AT4G09890
|
unknown |
-0.213 |
-0.743 |
| 435 |
264133_at |
AT1G79080
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.213 |
0.027 |
| 436 |
263130_at |
AT1G78650
|
POLD3 |
-0.212 |
-0.045 |
| 437 |
262624_at |
AT1G06450
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.212 |
0.284 |
| 438 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
-0.209 |
-0.136 |
| 439 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.209 |
-0.047 |
| 440 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
-0.209 |
0.401 |
| 441 |
251142_at |
AT5G01015
|
unknown |
-0.208 |
-0.418 |
| 442 |
251718_at |
AT3G56100
|
IMK3, MRLK, meristematic receptor-like kinase |
-0.208 |
-0.183 |
| 443 |
251800_at |
AT3G55510
|
RBL, REBELOTE |
-0.208 |
0.105 |
| 444 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.207 |
-0.291 |
| 445 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
-0.207 |
-0.244 |
| 446 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
-0.207 |
0.247 |
| 447 |
245723_at |
AT1G73400
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.207 |
0.032 |
| 448 |
266222_at |
AT2G28780
|
unknown |
-0.207 |
-0.557 |
| 449 |
260776_at |
AT1G14580
|
[C2H2-like zinc finger protein] |
-0.207 |
-0.274 |
| 450 |
258633_at |
AT3G07990
|
SCPL27, serine carboxypeptidase-like 27 |
-0.206 |
-0.091 |
| 451 |
249948_at |
AT5G19210
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.205 |
-0.222 |
| 452 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.205 |
-0.398 |
| 453 |
259199_at |
AT3G08980
|
[Peptidase S24/S26A/S26B/S26C family protein] |
-0.205 |
0.258 |
| 454 |
256010_at |
AT1G19220
|
ARF11, AUXIN RESPONSE FACTOR11, ARF19, auxin response factor 19, IAA22, indole-3-acetic acid inducible 22 |
-0.204 |
-0.357 |
| 455 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
-0.204 |
-0.554 |
| 456 |
264824_at |
AT1G03420
|
Sadhu4-2, sadhu non-coding retrotransposon 4-2 |
-0.204 |
-0.035 |
| 457 |
259358_at |
AT1G13250
|
GATL3, galacturonosyltransferase-like 3 |
-0.203 |
-0.415 |
| 458 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.203 |
-1.193 |
| 459 |
253583_at |
AT4G30680
|
[Initiation factor eIF-4 gamma, MA3] |
-0.202 |
0.069 |
| 460 |
264900_at |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
-0.202 |
-0.806 |
| 461 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.202 |
-0.624 |
| 462 |
267013_at |
AT2G39180
|
ATCRR2, CCR2, CRINKLY4 related 2 |
-0.201 |
-0.575 |
| 463 |
251058_at |
AT5G01790
|
unknown |
-0.201 |
-0.301 |
| 464 |
259891_at |
AT1G72730
|
[DEA(D/H)-box RNA helicase family protein] |
-0.200 |
0.089 |
| 465 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
-0.199 |
0.228 |
| 466 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-0.199 |
-0.447 |
| 467 |
262752_at |
AT1G16330
|
CYCB3;1, cyclin b3;1 |
-0.198 |
0.113 |
| 468 |
256091_at |
AT1G20693
|
HMG BETA 1, HIGH MOBILITY GROUP BETA 1, HMGB2, high mobility group B2, NFD02, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP D 02, NFD2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP D 2 |
-0.198 |
-0.222 |
| 469 |
256052_at |
AT1G06960
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.197 |
-0.109 |
| 470 |
263894_at |
AT2G21910
|
CYP96A5, cytochrome P450, family 96, subfamily A, polypeptide 5 |
-0.197 |
-0.105 |
| 471 |
257267_at |
AT3G15030
|
MEE35, maternal effect embryo arrest 35, TCP4, TCP family transcription factor 4 |
-0.197 |
-0.111 |
| 472 |
245533_at |
AT4G15130
|
ATCCT2, CCT2, phosphorylcholine cytidylyltransferase2 |
-0.196 |
0.052 |
| 473 |
267636_at |
AT2G42110
|
unknown |
-0.196 |
-0.168 |
| 474 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.196 |
0.103 |
| 475 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.195 |
-0.152 |
| 476 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.195 |
-0.298 |
| 477 |
253891_at |
AT4G27720
|
[Major facilitator superfamily protein] |
-0.194 |
-0.066 |
| 478 |
257071_at |
AT3G28180
|
ATCSLC04, Cellulose-synthase-like C4, ATCSLC4, CSLC04, CSLC04, Cellulose-synthase-like C4, CSLC4, CELLULOSE-SYNTHASE LIKE C4 |
-0.194 |
-0.143 |
| 479 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.194 |
0.101 |
| 480 |
251752_at |
AT3G55740
|
ATPROT2, PROLINE TRANSPORTER 2, PROT2, proline transporter 2 |
-0.193 |
-0.223 |
| 481 |
251852_at |
AT3G54750
|
unknown |
-0.193 |
0.506 |
| 482 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.193 |
-0.596 |
| 483 |
262136_at |
AT1G77850
|
ARF17, auxin response factor 17 |
-0.192 |
0.113 |
| 484 |
262951_at |
AT1G75500
|
UMAMIT5, Usually multiple acids move in and out Transporters 5, WAT1, Walls Are Thin 1 |
-0.192 |
-0.690 |
| 485 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.191 |
0.202 |
| 486 |
261230_at |
AT1G20010
|
TUB5, tubulin beta-5 chain |
-0.191 |
-0.369 |
| 487 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.191 |
-0.064 |
| 488 |
266262_at |
AT2G27590
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.190 |
0.066 |
| 489 |
245367_at |
AT4G16265
|
NRPB9B, NRPD9B, NRPE9B |
-0.190 |
0.140 |
| 490 |
252543_at |
AT3G45780
|
JK224, NPH1, NONPHOTOTROPIC HYPOCOTYL 1, PHOT1, phototropin 1, RPT1, ROOT PHOTOTROPISM 1 |
-0.190 |
-0.455 |
| 491 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-0.190 |
0.181 |
| 492 |
248177_at |
AT5G54630
|
[zinc finger protein-related] |
-0.190 |
-0.299 |
| 493 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
-0.190 |
0.201 |
| 494 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.190 |
-0.186 |
| 495 |
267580_at |
AT2G41990
|
unknown |
-0.189 |
-0.372 |
| 496 |
258242_at |
AT3G27640
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.189 |
0.680 |
| 497 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.188 |
-0.302 |