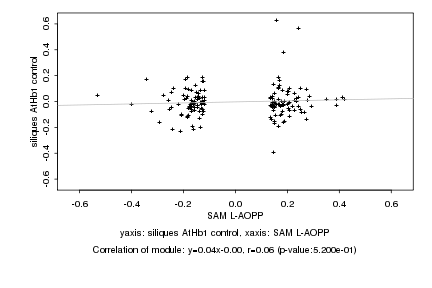
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
SAM L-AOPP |
Log2 signal ratio
siliques AtHb1 control |
| 1 |
252792_at |
AT3G42240
|
unknown |
0.418 |
0.017 |
| 2 |
260268_at |
AT1G68490
|
unknown |
0.409 |
0.037 |
| 3 |
254427_at |
AT4G21190
|
emb1417, embryo defective 1417 |
0.389 |
0.020 |
| 4 |
266753_at |
AT2G46990
|
IAA20, indole-3-acetic acid inducible 20 |
0.388 |
-0.026 |
| 5 |
246965_at |
AT5G24840
|
[tRNA (guanine-N-7) methyltransferase] |
0.350 |
0.020 |
| 6 |
248789_at |
AT5G47440
|
unknown |
0.292 |
-0.037 |
| 7 |
258423_at |
AT3G16730
|
HEB2, hypersensitive to excess boron 2 |
0.285 |
0.046 |
| 8 |
256254_at |
AT3G11290
|
unknown |
0.274 |
0.010 |
| 9 |
246020_at |
AT5G10710
|
[INVOLVED IN: chromosome segregation, cell division] |
0.273 |
0.094 |
| 10 |
262982_at |
AT1G54390
|
ING2, INHIBITOR OF GROWTH 2 |
0.272 |
-0.134 |
| 11 |
262921_at |
AT1G79430
|
APL, ALTERED PHLOEM DEVELOPMENT, WDY, WOODY |
0.263 |
-0.084 |
| 12 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.252 |
-0.084 |
| 13 |
253618_at |
AT4G30420
|
UMAMIT34, Usually multiple acids move in and out Transporters 34 |
0.248 |
-0.061 |
| 14 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
0.247 |
0.104 |
| 15 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.242 |
0.567 |
| 16 |
263642_at |
AT2G04750
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.241 |
0.034 |
| 17 |
264632_at |
AT1G65640
|
DEG4, degradation of periplasmic proteins 4, DegP4, DegP protease 4 |
0.239 |
-0.035 |
| 18 |
259307_at |
AT3G05230
|
[Signal peptidase subunit] |
0.235 |
0.005 |
| 19 |
252876_at |
AT4G39970
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.232 |
0.031 |
| 20 |
261102_at |
AT1G62880
|
[Cornichon family protein] |
0.226 |
-0.066 |
| 21 |
262553_at |
AT1G31360
|
ATRECQ2, ARABIDOPSIS THALIANA RECQ 2, MED34, RECQL2, RECQ helicase L2 |
0.225 |
0.065 |
| 22 |
256912_at |
AT3G23850
|
unknown |
0.224 |
0.011 |
| 23 |
267368_at |
AT2G44350
|
ATCS, CSY4, CITRATE SYNTHASE 4 |
0.217 |
-0.033 |
| 24 |
248432_at |
AT5G51390
|
unknown |
0.208 |
0.102 |
| 25 |
249505_at |
AT5G38870
|
[pseudogene] |
0.207 |
-0.062 |
| 26 |
266638_at |
AT2G35490
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
0.207 |
-0.006 |
| 27 |
259711_at |
AT1G77570
|
AtREN1, REN1, restricted to nucleolus 1 |
0.205 |
-0.109 |
| 28 |
250439_at |
AT5G10450
|
14-3-3lambda, 14-3-3 PROTEIN G-BOX FACTOR14 LAMBDA, AFT1, GRF6, G-box regulating factor 6 |
0.203 |
-0.015 |
| 29 |
247686_at |
AT5G59700
|
[Protein kinase superfamily protein] |
0.202 |
0.084 |
| 30 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.201 |
-0.049 |
| 31 |
266618_at |
AT2G35480
|
unknown |
0.199 |
0.058 |
| 32 |
262696_at |
AT1G75870
|
unknown |
0.198 |
-0.021 |
| 33 |
258282_at |
AT3G26910
|
[hydroxyproline-rich glycoprotein family protein] |
0.198 |
0.083 |
| 34 |
248042_at |
AT5G55960
|
unknown |
0.186 |
-0.153 |
| 35 |
264643_at |
AT1G08990
|
GUX5, Glucuronic Acid Substitution of Xylan 5, PGSIP5, plant glycogenin-like starch initiation protein 5 |
0.186 |
0.006 |
| 36 |
264890_at |
AT1G23180
|
[ARM repeat superfamily protein] |
0.185 |
-0.025 |
| 37 |
254574_at |
AT4G19430
|
unknown |
0.184 |
-0.160 |
| 38 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
0.182 |
0.385 |
| 39 |
260538_at |
AT2G43460
|
[Ribosomal L38e protein family] |
0.182 |
-0.018 |
| 40 |
254400_at |
AT4G21270
|
ATK1, kinesin 1, KATA, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA A, KATAP, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA A PROTEIN |
0.181 |
0.091 |
| 41 |
259124_at |
AT3G02310
|
AGL4, AGAMOUS-like 4, SEP2, SEPALLATA 2 |
0.179 |
-0.026 |
| 42 |
259889_at |
AT1G76405
|
unknown |
0.178 |
-0.016 |
| 43 |
247575_at |
AT5G61030
|
GR-RBP3, glycine-rich RNA-binding protein 3 |
0.178 |
-0.070 |
| 44 |
261279_at |
AT1G05850
|
ATCTL1, CTL1, CHITINASE-LIKE PROTEIN 1, ELP, ECTOPIC DEPOSITION OF LIGNIN IN PITH, ELP1, ECTOPIC DEPOSITION OF LIGNIN IN PITH 1, ERH2, ECTOPIC ROOT HAIR 2, HOT2, SENSITIVE TO HOT TEMPERATURES 2, POM1, POM-POM1 |
0.175 |
-0.094 |
| 45 |
247870_at |
AT5G57580
|
[Calmodulin-binding protein] |
0.174 |
0.001 |
| 46 |
253477_at |
AT4G32320
|
APX6, ascorbate peroxidase 6 |
0.173 |
-0.104 |
| 47 |
259528_at |
AT1G12330
|
unknown |
0.171 |
-0.037 |
| 48 |
248072_at |
AT5G55680
|
[glycine-rich protein] |
0.169 |
-0.016 |
| 49 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.169 |
0.127 |
| 50 |
262373_at |
AT1G73120
|
unknown |
0.167 |
0.168 |
| 51 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.165 |
0.187 |
| 52 |
250784_at |
AT5G05480
|
[Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A protein] |
0.165 |
0.107 |
| 53 |
262648_at |
AT1G14030
|
LSMT-L, lysine methyltransferase (LSMT)-like |
0.165 |
0.018 |
| 54 |
256922_at |
AT3G19010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.164 |
0.116 |
| 55 |
250085_at |
AT5G17250
|
[Alkaline-phosphatase-like family protein] |
0.163 |
-0.019 |
| 56 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.162 |
-0.188 |
| 57 |
260092_at |
AT1G73280
|
scpl3, serine carboxypeptidase-like 3 |
0.158 |
-0.017 |
| 58 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.157 |
0.634 |
| 59 |
248952_at |
AT5G45410
|
unknown |
0.154 |
-0.101 |
| 60 |
245175_at |
AT2G47470
|
ATPDI11, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 11, ATPDIL2-1, PDI-LIKE 2-1, MEE30, MATERNAL EFFECT EMBRYO ARREST 30, PDI11, PROTEIN DISULFIDE ISOMERASE 11, UNE5, UNFERTILIZED EMBRYO SAC 5 |
0.152 |
-0.013 |
| 61 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
0.150 |
-0.040 |
| 62 |
248479_at |
AT5G50910
|
unknown |
0.149 |
0.003 |
| 63 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
0.149 |
-0.045 |
| 64 |
245594_at |
AT4G14570
|
AARE, acylamino acid-releasing enzyme, AtAARE |
0.149 |
-0.154 |
| 65 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
0.148 |
-0.163 |
| 66 |
256920_at |
AT3G18980
|
ETP1, EIN2 targeting protein1 |
0.147 |
-0.036 |
| 67 |
246450_at |
AT5G16820
|
ATHSF3, ARABIDOPSIS HEAT SHOCK FACTOR 3, ATHSFA1B, ARABIDOPSIS THALIANA CLASS A HEAT SHOCK FACTOR 1B, HSF3, heat shock factor 3, HSFA1B, CLASS A HEAT SHOCK FACTOR 1B |
0.147 |
0.063 |
| 68 |
258612_at |
AT3G02920
|
ATRPA32B, RPA32B |
0.146 |
-0.068 |
| 69 |
259189_at |
AT3G01700
|
AGP11, arabinogalactan protein 11, ATAGP11, ARABINOGALACTAN PROTEIN 11 |
0.146 |
-0.392 |
| 70 |
264633_at |
AT1G65660
|
SMP1, SWELLMAP 1 |
0.144 |
0.134 |
| 71 |
257996_at |
AT3G19950
|
[RING/U-box superfamily protein] |
0.143 |
-0.026 |
| 72 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
0.141 |
-0.004 |
| 73 |
264306_at |
AT1G78890
|
unknown |
0.141 |
-0.042 |
| 74 |
246548_at |
AT5G14910
|
[Heavy metal transport/detoxification superfamily protein ] |
0.141 |
-0.016 |
| 75 |
245430_at |
AT4G17060
|
FIP2, FRIGIDA interacting protein 2 |
0.140 |
0.046 |
| 76 |
245283_at |
AT4G13980
|
AT-HSFA5, HSFA5, HEAT SHOCK TRANSCRIPTION FACTOR A5 |
0.139 |
0.025 |
| 77 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
0.137 |
-0.136 |
| 78 |
251739_at |
AT3G56170
|
CAN, Ca-2+ dependent nuclease, CAN1, calcium dependent nuclease 1 |
0.136 |
-0.027 |
| 79 |
263527_at |
AT2G24810
|
[Pathogenesis-related thaumatin superfamily protein] |
0.135 |
-0.038 |
| 80 |
247438_at |
AT5G62460
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.134 |
-0.116 |
| 81 |
262388_at |
AT1G49320
|
unknown |
0.134 |
0.032 |
| 82 |
248705_at |
AT5G48520
|
AtAUG3, AUG3, augmin 3 |
0.134 |
0.024 |
| 83 |
262568_at |
AT1G34290
|
AtRLP5, receptor like protein 5, RLP5, receptor like protein 5 |
0.134 |
-0.029 |
| 84 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.532 |
0.052 |
| 85 |
245723_at |
AT1G73400
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.402 |
-0.022 |
| 86 |
263363_at |
AT2G03850
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.344 |
0.173 |
| 87 |
250969_at |
AT5G02740
|
[Ribosomal protein S24e family protein] |
-0.324 |
-0.076 |
| 88 |
245527_at |
AT4G15955
|
[alpha/beta-Hydrolases superfamily protein] |
-0.295 |
-0.161 |
| 89 |
256820_at |
AT3G22170
|
CPD45, CHLOROPLAST DIVISION MUTANT 45, FHY3, far-red elongated hypocotyls 3 |
-0.277 |
0.052 |
| 90 |
248250_at |
AT5G53130
|
ATCNGC1, CYCLIC NUCLEOTIDE-GATED CHANNEL 1, CNGC1, cyclic nucleotide gated channel 1 |
-0.261 |
0.013 |
| 91 |
259714_at |
AT1G60980
|
ATGA20OX4, gibberellin 20-oxidase 4, GA20OX4, gibberellin 20-oxidase 4 |
-0.256 |
-0.060 |
| 92 |
266153_at |
AT2G12320
|
unknown |
-0.248 |
-0.039 |
| 93 |
248370_at |
AT5G52170
|
HDG7, homeodomain GLABROUS 7 |
-0.248 |
0.071 |
| 94 |
259713_at |
AT1G77610
|
[EamA-like transporter family protein] |
-0.243 |
-0.215 |
| 95 |
251010_at |
AT5G02550
|
unknown |
-0.242 |
0.105 |
| 96 |
252595_at |
AT3G45690
|
[Major facilitator superfamily protein] |
-0.221 |
-0.018 |
| 97 |
256323_at |
AT1G54920
|
unknown |
-0.213 |
-0.229 |
| 98 |
257995_at |
AT3G19940
|
[Major facilitator superfamily protein] |
-0.211 |
-0.093 |
| 99 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
-0.209 |
-0.105 |
| 100 |
246039_at |
AT5G19480
|
unknown |
-0.203 |
0.051 |
| 101 |
258120_at |
AT3G14730
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.199 |
0.017 |
| 102 |
247565_at |
AT5G61150
|
VIP4, VERNALIZATION INDEPENDENCE 4 |
-0.196 |
0.173 |
| 103 |
251022_at |
AT5G02150
|
Fes1C, Fes1C |
-0.196 |
0.106 |
| 104 |
252431_at |
AT3G47700
|
MAG2, maigo2 |
-0.192 |
0.031 |
| 105 |
258619_at |
AT3G02780
|
IDI2, ATISOPENTENYL DIPHOSPHE ISOMERASE 2, IPIAT1, IPP2, isopentenyl pyrophosphate:dimethylallyl pyrophosphate isomerase 2 |
-0.187 |
0.041 |
| 106 |
250358_at |
AT5G11740
|
AGP15, arabinogalactan protein 15, ATAGP15 |
-0.187 |
-0.113 |
| 107 |
260161_at |
AT1G79860
|
ATROPGEF12, MEE64, MATERNAL EFFECT EMBRYO ARREST 64, ROPGEF12, ROP (rho of plants) guanine nucleotide exchange factor 12 |
-0.186 |
-0.121 |
| 108 |
261526_at |
AT1G14370
|
APK2A, protein kinase 2A, Kin1, kinase 1, PBL2, PBS1-like 2 |
-0.186 |
0.192 |
| 109 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
-0.183 |
-0.052 |
| 110 |
250143_at |
AT5G14670
|
ARFA1B, ADP-ribosylation factor A1B, ATARFA1B, ADP-ribosylation factor A1B |
-0.182 |
-0.103 |
| 111 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
-0.182 |
0.098 |
| 112 |
260178_at |
AT1G70720
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.179 |
-0.031 |
| 113 |
257562_at |
AT3G30480
|
unknown |
-0.177 |
-0.040 |
| 114 |
248199_at |
AT5G54170
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.175 |
-0.039 |
| 115 |
252762_at |
AT3G42780
|
unknown |
-0.175 |
-0.022 |
| 116 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
-0.173 |
0.089 |
| 117 |
254254_at |
AT4G23330
|
unknown |
-0.173 |
-0.054 |
| 118 |
265250_at |
AT2G01950
|
BRL2, BRI1-like 2, VH1, VASCULAR HIGHWAY 1 |
-0.172 |
-0.013 |
| 119 |
251858_at |
AT3G54820
|
PIP2;5, plasma membrane intrinsic protein 2;5, PIP2D, PLASMA MEMBRANE INTRINSIC PROTEIN 2D |
-0.171 |
-0.075 |
| 120 |
251341_at |
AT3G60770
|
[Ribosomal protein S13/S15] |
-0.171 |
-0.033 |
| 121 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
-0.171 |
-0.020 |
| 122 |
252541_at |
AT3G45750
|
[Nucleotidyltransferase family protein] |
-0.170 |
-0.037 |
| 123 |
258839_at |
AT3G07170
|
[Sterile alpha motif (SAM) domain-containing protein] |
-0.170 |
0.009 |
| 124 |
261524_at |
AT1G14300
|
[ARM repeat superfamily protein] |
-0.168 |
-0.191 |
| 125 |
262142_at |
AT1G52640
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.166 |
-0.005 |
| 126 |
257361_at |
AT2G07740
|
[similar to nucleic acid binding / zinc ion binding [Arabidopsis thaliana] (TAIR:AT2G01050.1)] |
-0.164 |
-0.019 |
| 127 |
249251_at |
AT5G42000
|
[ORMDL family protein] |
-0.163 |
-0.210 |
| 128 |
257258_at |
AT3G22040
|
unknown |
-0.161 |
-0.011 |
| 129 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.160 |
-0.068 |
| 130 |
257087_at |
AT3G20500
|
ATPAP18, PURPLE ACID PHOSPHATASE 18, PAP18, purple acid phosphatase 18 |
-0.159 |
0.001 |
| 131 |
254782_at |
AT4G12860
|
UNE14, unfertilized embryo sac 14 |
-0.159 |
-0.041 |
| 132 |
246064_at |
AT5G19350
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.156 |
0.127 |
| 133 |
250607_at |
AT5G07370
|
ATIPK2A, INOSITOL POLYPHOSPHATE KINASE 2 ALPHA, IMPK, inositol multi-phosphate kinase, IPK2a, inositol polyphosphate kinase 2 alpha |
-0.156 |
0.032 |
| 134 |
266197_at |
AT2G39120
|
WTF9, what's this factor 9 |
-0.153 |
0.072 |
| 135 |
262885_at |
AT1G64740
|
TUA1, alpha-1 tubulin |
-0.149 |
-0.040 |
| 136 |
248914_at |
AT5G45750
|
AtRABA1c, RAB GTPase homolog A1C, RABA1c, RAB GTPase homolog A1C |
-0.149 |
0.037 |
| 137 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.148 |
-0.029 |
| 138 |
245092_at |
AT2G40950
|
BZIP17 |
-0.148 |
0.019 |
| 139 |
260472_at |
AT1G10990
|
unknown |
-0.147 |
-0.029 |
| 140 |
261250_at |
AT1G05890
|
ARI5, ARIADNE 5, ATARI5, ARABIDOPSIS ARIADNE 5 |
-0.145 |
0.040 |
| 141 |
252264_at |
AT3G49590
|
ATG13, autophagy-related 13 |
-0.144 |
0.063 |
| 142 |
259780_at |
AT1G29630
|
[5'-3' exonuclease family protein] |
-0.141 |
-0.070 |
| 143 |
248354_at |
AT5G52330
|
[TRAF-like superfamily protein] |
-0.139 |
0.026 |
| 144 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.139 |
-0.125 |
| 145 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.137 |
-0.003 |
| 146 |
261999_at |
AT1G33800
|
AtGXMT1, GXM1, glucuronoxylan methyltransferase 1, GXMT1, glucuronoxylan methyltransferase 1 |
-0.137 |
-0.200 |
| 147 |
260067_at |
AT1G73780
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.136 |
0.087 |
| 148 |
263589_at |
AT2G25280
|
unknown |
-0.134 |
-0.051 |
| 149 |
261241_at |
AT1G32950
|
[Subtilase family protein] |
-0.132 |
-0.004 |
| 150 |
261162_at |
AT1G34440
|
unknown |
-0.131 |
-0.018 |
| 151 |
246907_at |
AT5G25510
|
[Protein phosphatase 2A regulatory B subunit family protein] |
-0.130 |
-0.059 |
| 152 |
256113_at |
AT1G16810
|
unknown |
-0.130 |
0.035 |
| 153 |
253863_at |
AT4G27420
|
ABCG9, ATP-binding cassette G9 |
-0.129 |
0.004 |
| 154 |
252142_at |
AT3G51120
|
[DNA binding] |
-0.128 |
0.161 |
| 155 |
249990_at |
AT5G18540
|
unknown |
-0.128 |
0.187 |
| 156 |
250081_at |
AT5G17180
|
unknown |
-0.127 |
-0.004 |
| 157 |
262674_at |
AT1G75910
|
EXL4, extracellular lipase 4 |
-0.127 |
-0.099 |
| 158 |
251444_at |
AT3G59990
|
MAP2B, methionine aminopeptidase 2B |
-0.127 |
-0.055 |
| 159 |
247481_at |
AT5G62400
|
unknown |
-0.125 |
-0.013 |
| 160 |
246303_at |
AT3G51870
|
[Mitochondrial substrate carrier family protein] |
-0.125 |
-0.073 |
| 161 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
-0.123 |
0.159 |
| 162 |
245142_at |
AT2G45270
|
GCP1, glycoprotease 1 |
-0.122 |
0.014 |
| 163 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
-0.122 |
-0.023 |
| 164 |
257855_at |
AT3G13040
|
[myb-like HTH transcriptional regulator family protein] |
-0.122 |
0.092 |
| 165 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
-0.121 |
0.038 |