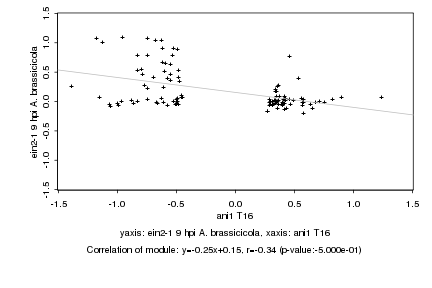
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ani1 T16 |
Log2 signal ratio
ein2-1 9 hpi A. brassicicola |
| 1 |
256382_at |
AT1G66860
|
[Class I glutamine amidotransferase-like superfamily protein] |
1.234 |
0.077 |
| 2 |
249932_at |
AT5G22390
|
unknown |
0.893 |
0.085 |
| 3 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
0.815 |
0.041 |
| 4 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.753 |
-0.016 |
| 5 |
266454_at |
AT2G22750
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.712 |
0.001 |
| 6 |
251481_at |
AT3G59730
|
LecRK-V.6, L-type lectin receptor kinase V.6 |
0.673 |
-0.008 |
| 7 |
263120_at |
AT1G78490
|
CYP708A3, cytochrome P450, family 708, subfamily A, polypeptide 3 |
0.645 |
-0.113 |
| 8 |
247800_at |
AT5G58570
|
unknown |
0.631 |
-0.043 |
| 9 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
0.576 |
-0.004 |
| 10 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
0.576 |
-0.197 |
| 11 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.571 |
0.044 |
| 12 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
0.563 |
-0.015 |
| 13 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.561 |
-0.059 |
| 14 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
0.560 |
0.063 |
| 15 |
251293_at |
AT3G61930
|
unknown |
0.532 |
0.406 |
| 16 |
251480_at |
AT3G59710
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.486 |
0.023 |
| 17 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.463 |
-0.049 |
| 18 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
0.457 |
0.041 |
| 19 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.451 |
0.778 |
| 20 |
255008_at |
AT4G10060
|
[Beta-glucosidase, GBA2 type family protein] |
0.425 |
-0.102 |
| 21 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.422 |
0.052 |
| 22 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.420 |
0.028 |
| 23 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.411 |
-0.128 |
| 24 |
262659_at |
AT1G14240
|
[GDA1/CD39 nucleoside phosphatase family protein] |
0.409 |
-0.026 |
| 25 |
265452_at |
AT2G46510
|
AIB, ABA-inducible BHLH-type transcription factor, ATAIB, ABA-inducible BHLH-type transcription factor, JAM1, JA-associated MYC2-like 1 |
0.409 |
0.090 |
| 26 |
247772_at |
AT5G58610
|
[PHD finger transcription factor, putative] |
0.406 |
-0.033 |
| 27 |
245728_at |
AT1G73340
|
[Cytochrome P450 superfamily protein] |
0.406 |
0.035 |
| 28 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.396 |
-0.054 |
| 29 |
258874_at |
AT3G03230
|
[alpha/beta-Hydrolases superfamily protein] |
0.392 |
-0.011 |
| 30 |
247536_at |
AT5G61650
|
CYCP4, CYCP4;2, CYCLIN P4;2 |
0.387 |
-0.035 |
| 31 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.371 |
0.099 |
| 32 |
260090_at |
AT1G73310
|
scpl4, serine carboxypeptidase-like 4 |
0.365 |
-0.050 |
| 33 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.362 |
0.032 |
| 34 |
255884_at |
AT1G20310
|
unknown |
0.360 |
0.283 |
| 35 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
0.355 |
-0.106 |
| 36 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.354 |
0.267 |
| 37 |
262043_at |
AT1G80190
|
PSF1, partner of SLD five 1 |
0.351 |
0.016 |
| 38 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
0.346 |
0.101 |
| 39 |
249108_at |
AT5G43690
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.344 |
0.016 |
| 40 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.341 |
0.187 |
| 41 |
253070_at |
AT4G37850
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.340 |
-0.013 |
| 42 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.337 |
-0.028 |
| 43 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.336 |
0.183 |
| 44 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
0.336 |
0.220 |
| 45 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
0.334 |
0.018 |
| 46 |
255931_at |
AT1G12710
|
AtPP2-A12, phloem protein 2-A12, PP2-A12, phloem protein 2-A12 |
0.331 |
0.020 |
| 47 |
248723_at |
AT5G47950
|
[HXXXD-type acyl-transferase family protein] |
0.318 |
-0.048 |
| 48 |
247581_at |
AT5G61350
|
[Protein kinase superfamily protein] |
0.312 |
-0.065 |
| 49 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.311 |
0.013 |
| 50 |
245141_at |
AT2G45400
|
BEN1, BRI1-5 ENHANCED 1 |
0.309 |
-0.047 |
| 51 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.295 |
-0.039 |
| 52 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
0.291 |
0.006 |
| 53 |
255626_at |
AT4G00780
|
[TRAF-like family protein] |
0.287 |
-0.067 |
| 54 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
0.283 |
0.051 |
| 55 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.281 |
-0.013 |
| 56 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
0.270 |
-0.169 |
| 57 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.267 |
-0.166 |
| 58 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
-1.396 |
0.257 |
| 59 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-1.179 |
1.073 |
| 60 |
251755_at |
AT3G55790
|
unknown |
-1.156 |
0.078 |
| 61 |
245082_at |
AT2G23270
|
unknown |
-1.132 |
1.009 |
| 62 |
267573_at |
AT2G30670
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.069 |
-0.041 |
| 63 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
-1.065 |
-0.076 |
| 64 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
-1.006 |
-0.019 |
| 65 |
245875_at |
AT1G26240
|
[Proline-rich extensin-like family protein] |
-0.998 |
-0.055 |
| 66 |
259558_at |
AT1G21230
|
WAK5, wall associated kinase 5 |
-0.971 |
0.002 |
| 67 |
253796_at |
AT4G28460
|
unknown |
-0.962 |
1.095 |
| 68 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
-0.888 |
0.027 |
| 69 |
246333_at |
AT3G44840
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.868 |
-0.022 |
| 70 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.835 |
0.003 |
| 71 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.834 |
0.787 |
| 72 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.832 |
0.541 |
| 73 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.801 |
0.561 |
| 74 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
-0.790 |
0.463 |
| 75 |
259197_at |
AT3G03670
|
[Peroxidase superfamily protein] |
-0.776 |
0.275 |
| 76 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
-0.754 |
0.788 |
| 77 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
-0.754 |
1.075 |
| 78 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
-0.753 |
0.036 |
| 79 |
264314_at |
AT1G70420
|
unknown |
-0.750 |
0.222 |
| 80 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
-0.700 |
0.416 |
| 81 |
246370_at |
AT1G51920
|
unknown |
-0.678 |
1.039 |
| 82 |
265063_at |
AT1G61500
|
[S-locus lectin protein kinase family protein] |
-0.672 |
-0.016 |
| 83 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.665 |
-0.022 |
| 84 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
-0.635 |
1.042 |
| 85 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
-0.632 |
0.061 |
| 86 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
-0.625 |
0.666 |
| 87 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.623 |
0.912 |
| 88 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
-0.617 |
0.244 |
| 89 |
267572_at |
AT2G30660
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
-0.611 |
-0.000 |
| 90 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
-0.604 |
0.515 |
| 91 |
250796_at |
AT5G05300
|
unknown |
-0.597 |
0.657 |
| 92 |
261221_at |
AT1G19960
|
unknown |
-0.583 |
-0.058 |
| 93 |
264774_at |
AT1G22890
|
unknown |
-0.583 |
0.405 |
| 94 |
257927_at |
AT3G23240
|
ATERF1, ETHYLENE RESPONSE FACTOR 1, ERF1, ethylene response factor 1 |
-0.557 |
0.366 |
| 95 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
-0.556 |
0.467 |
| 96 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-0.556 |
0.647 |
| 97 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
-0.542 |
0.784 |
| 98 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-0.530 |
0.907 |
| 99 |
254178_at |
AT4G23880
|
unknown |
-0.528 |
0.001 |
| 100 |
245692_at |
AT5G04150
|
BHLH101 |
-0.513 |
-0.044 |
| 101 |
246860_at |
AT5G25840
|
unknown |
-0.505 |
-0.031 |
| 102 |
259316_at |
AT3G01175
|
unknown |
-0.505 |
-0.036 |
| 103 |
264759_at |
AT1G61480
|
[S-locus lectin protein kinase family protein] |
-0.503 |
0.037 |
| 104 |
248570_at |
AT5G49780
|
[Leucine-rich repeat protein kinase family protein] |
-0.494 |
0.059 |
| 105 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
-0.492 |
0.889 |
| 106 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
-0.492 |
0.001 |
| 107 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.490 |
-0.034 |
| 108 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
-0.490 |
0.529 |
| 109 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
-0.487 |
0.417 |
| 110 |
261249_at |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
-0.479 |
0.341 |
| 111 |
252569_at |
AT3G45420
|
LecRK-I.4, L-type lectin receptor kinase I.4 |
-0.462 |
0.073 |
| 112 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
-0.460 |
0.118 |
| 113 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.456 |
0.078 |