|
probeID |
AGICode |
Annotation |
Log2 signal ratio
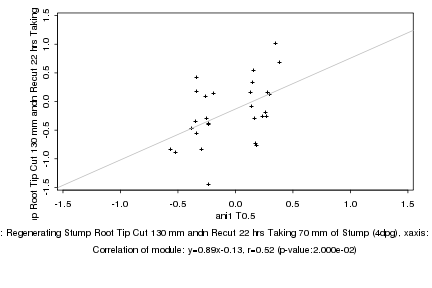
ani1 T0.5 |
Log2 signal ratio
Regenerating Stump Root Tip Cut 130 mm andn Recut 22 hrs Taking 70 mm of Stump (4dpg) |
| 1 |
254187_at |
AT4G23890
|
CRR31, CHLORORESPIRATORY REDUCTION 31, NdhS, NADH dehydrogenase-like complex S |
0.379 |
0.693 |
| 2 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.348 |
1.021 |
| 3 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
0.288 |
0.139 |
| 4 |
251658_at |
AT3G57020
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.273 |
0.160 |
| 5 |
246510_at |
AT5G15410
|
ATCNGC2, CYCLIC NUCLEOTIDE-GATED CHANNEL 2, CNGC2, CYCLIC NUCLEOTIDE GATED CHANNEL 2, DND1, DEFENSE NO DEATH 1 |
0.265 |
-0.259 |
| 6 |
245126_at |
AT2G47460
|
ATMYB12, MYB DOMAIN PROTEIN 12, MYB12, myb domain protein 12, PFG1, PRODUCTION OF FLAVONOL GLYCOSIDES 1 |
0.254 |
-0.180 |
| 7 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.235 |
-0.249 |
| 8 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
0.183 |
-0.755 |
| 9 |
252092_at |
AT3G51420
|
ATSSL4, STRICTOSIDINE SYNTHASE-LIKE 4, SSL4, strictosidine synthase-like 4 |
0.174 |
-0.732 |
| 10 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
0.163 |
-0.296 |
| 11 |
257172_at |
AT3G23700
|
[Nucleic acid-binding proteins superfamily] |
0.156 |
0.551 |
| 12 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
0.148 |
0.346 |
| 13 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.138 |
-0.071 |
| 14 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
0.128 |
0.173 |
| 15 |
266392_at |
AT2G41280
|
ATM10, M10 |
-0.572 |
-0.824 |
| 16 |
246349_at |
AT1G51915
|
[cryptdin protein-related] |
-0.524 |
-0.877 |
| 17 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
-0.389 |
-0.467 |
| 18 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
-0.349 |
-0.345 |
| 19 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.344 |
0.189 |
| 20 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.342 |
-0.552 |
| 21 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
-0.340 |
0.430 |
| 22 |
245662_at |
AT1G28190
|
unknown |
-0.296 |
-0.822 |
| 23 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
-0.264 |
0.099 |
| 24 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.256 |
-0.296 |
| 25 |
252596_at |
AT3G45330
|
LecRK-I.1, L-type lectin receptor kinase I.1 |
-0.241 |
-0.384 |
| 26 |
249896_at |
AT5G22530
|
unknown |
-0.239 |
-0.383 |
| 27 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-0.238 |
-1.432 |
| 28 |
253643_at |
AT4G29780
|
unknown |
-0.198 |
0.153 |