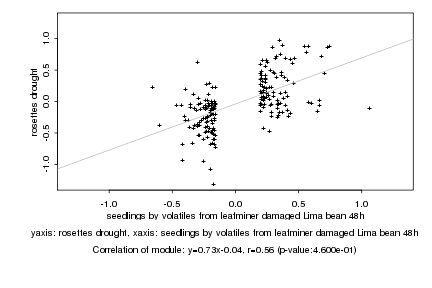
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
seedlings by volatiles from leafminer damaged Lima bean 48h |
Log2 signal ratio
rosettes drought |
| 1 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
1.057 |
-0.106 |
| 2 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
0.744 |
0.880 |
| 3 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.722 |
0.868 |
| 4 |
260561_at |
AT2G43580
|
[Chitinase family protein] |
0.701 |
0.456 |
| 5 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.678 |
0.728 |
| 6 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.665 |
-0.053 |
| 7 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
0.658 |
0.020 |
| 8 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.643 |
-0.153 |
| 9 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.598 |
-0.018 |
| 10 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.578 |
-0.000 |
| 11 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.572 |
0.879 |
| 12 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.555 |
0.787 |
| 13 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.546 |
0.882 |
| 14 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.461 |
0.688 |
| 15 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.457 |
0.298 |
| 16 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.444 |
0.608 |
| 17 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.432 |
0.679 |
| 18 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.430 |
-0.188 |
| 19 |
256053_at |
AT1G07260
|
UGT71C3, UDP-glucosyl transferase 71C3 |
0.419 |
-0.225 |
| 20 |
253155_at |
AT4G35720
|
unknown |
0.410 |
0.087 |
| 21 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.408 |
0.338 |
| 22 |
245882_at |
AT5G09470
|
DIC3, dicarboxylate carrier 3 |
0.408 |
-0.133 |
| 23 |
267646_at |
AT2G32830
|
PHT1;5, phosphate transporter 1;5, PHT5, PHOSPHATE TRANSPORTER 5 |
0.395 |
0.152 |
| 24 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.392 |
0.691 |
| 25 |
259552_at |
AT1G21320
|
[nucleotide binding] |
0.391 |
-0.057 |
| 26 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.384 |
0.388 |
| 27 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
0.377 |
0.049 |
| 28 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.372 |
0.891 |
| 29 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
0.370 |
-0.168 |
| 30 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.360 |
0.463 |
| 31 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.357 |
0.425 |
| 32 |
258646_at |
AT3G08040
|
ATFRD3, FRD3, FERRIC REDUCTASE DEFECTIVE 3, MAN1, MANGANESE ACCUMULATOR 1 |
0.353 |
0.138 |
| 33 |
252888_at |
AT4G39210
|
APL3 |
0.350 |
0.751 |
| 34 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
0.349 |
-0.025 |
| 35 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.346 |
0.979 |
| 36 |
266139_at |
AT2G28085
|
SAUR42, SMALL AUXIN UPREGULATED RNA 42 |
0.341 |
-0.160 |
| 37 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.340 |
-0.024 |
| 38 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
0.338 |
-0.218 |
| 39 |
260941_at |
AT1G44970
|
[Peroxidase superfamily protein] |
0.334 |
-0.033 |
| 40 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.332 |
0.025 |
| 41 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.332 |
-0.107 |
| 42 |
256509_at |
AT1G75300
|
[NmrA-like negative transcriptional regulator family protein] |
0.329 |
-0.039 |
| 43 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.329 |
-0.240 |
| 44 |
245352_at |
AT4G15490
|
UGT84A3 |
0.322 |
0.393 |
| 45 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.320 |
0.722 |
| 46 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.310 |
0.685 |
| 47 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.302 |
0.457 |
| 48 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
0.300 |
0.087 |
| 49 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
0.300 |
0.144 |
| 50 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.297 |
0.475 |
| 51 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.286 |
0.871 |
| 52 |
254759_at |
AT4G13180
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.283 |
0.223 |
| 53 |
257643_at |
AT3G25730
|
EDF3, ethylene response DNA binding factor 3 |
0.279 |
-0.047 |
| 54 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.279 |
-0.227 |
| 55 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
0.278 |
-0.164 |
| 56 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.277 |
-0.060 |
| 57 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.274 |
0.500 |
| 58 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.267 |
0.039 |
| 59 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.265 |
0.230 |
| 60 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.264 |
-0.474 |
| 61 |
248271_at |
AT5G53420
|
[CCT motif family protein] |
0.257 |
0.096 |
| 62 |
256924_at |
AT3G29590
|
AT5MAT |
0.252 |
0.627 |
| 63 |
266296_at |
AT2G29420
|
ATGSTU7, glutathione S-transferase tau 7, GST25, GLUTATHIONE S-TRANSFERASE 25, GSTU7, glutathione S-transferase tau 7 |
0.245 |
0.140 |
| 64 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.244 |
0.654 |
| 65 |
260639_at |
AT1G53180
|
unknown |
0.243 |
0.213 |
| 66 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
0.240 |
0.158 |
| 67 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
0.239 |
0.079 |
| 68 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.238 |
0.366 |
| 69 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.236 |
0.563 |
| 70 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
0.236 |
0.429 |
| 71 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.235 |
0.305 |
| 72 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
0.232 |
0.372 |
| 73 |
255230_at |
AT4G05390
|
ATRFNR1, root FNR 1, RFNR1, root FNR 1 |
0.229 |
-0.041 |
| 74 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
0.227 |
0.054 |
| 75 |
255056_at |
AT4G09820
|
AtTT8, BHLH42, TT8, TRANSPARENT TESTA 8 |
0.227 |
0.233 |
| 76 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.221 |
-0.425 |
| 77 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
0.220 |
0.156 |
| 78 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.220 |
0.177 |
| 79 |
261607_at |
AT1G49660
|
AtCXE5, carboxyesterase 5, CXE5, carboxyesterase 5 |
0.218 |
0.088 |
| 80 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.216 |
-0.084 |
| 81 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.216 |
0.032 |
| 82 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.214 |
0.244 |
| 83 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.210 |
0.320 |
| 84 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
0.210 |
0.250 |
| 85 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.208 |
0.663 |
| 86 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.207 |
0.252 |
| 87 |
251725_at |
AT3G56260
|
unknown |
0.205 |
0.420 |
| 88 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
0.204 |
0.040 |
| 89 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.203 |
0.076 |
| 90 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
0.203 |
0.128 |
| 91 |
253097_at |
AT4G37320
|
CYP81D5, cytochrome P450, family 81, subfamily D, polypeptide 5 |
0.202 |
0.378 |
| 92 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
0.202 |
-0.042 |
| 93 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.201 |
0.480 |
| 94 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
0.198 |
-0.154 |
| 95 |
247488_at |
AT5G61820
|
unknown |
0.196 |
0.455 |
| 96 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.196 |
0.163 |
| 97 |
261125_at |
AT1G04990
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.195 |
0.271 |
| 98 |
248820_at |
AT5G47060
|
unknown |
0.194 |
0.594 |
| 99 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
0.194 |
-0.027 |
| 100 |
258367_at |
AT3G14370
|
WAG2 |
0.193 |
-0.057 |
| 101 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
0.192 |
0.365 |
| 102 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.659 |
0.236 |
| 103 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.604 |
-0.367 |
| 104 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.471 |
-0.062 |
| 105 |
256066_at |
AT1G06980
|
unknown |
-0.430 |
-0.055 |
| 106 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.422 |
-0.674 |
| 107 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.421 |
-0.928 |
| 108 |
254954_at |
AT4G10910
|
unknown |
-0.404 |
-0.226 |
| 109 |
256096_at |
AT1G13650
|
unknown |
-0.399 |
-0.297 |
| 110 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-0.397 |
0.204 |
| 111 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.374 |
-0.300 |
| 112 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.366 |
-0.400 |
| 113 |
244934_at |
ATCG01080
|
NDHG |
-0.362 |
-0.081 |
| 114 |
244962_at |
ATCG01050
|
NDHD |
-0.361 |
-0.040 |
| 115 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.343 |
-0.659 |
| 116 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.337 |
-0.417 |
| 117 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
-0.334 |
0.122 |
| 118 |
244933_at |
ATCG01070
|
NDHE |
-0.329 |
-0.109 |
| 119 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.328 |
-0.376 |
| 120 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
-0.315 |
-0.131 |
| 121 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.306 |
-0.351 |
| 122 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.305 |
-0.396 |
| 123 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
-0.302 |
0.622 |
| 124 |
244932_at |
ATCG01060
|
PSAC |
-0.300 |
-0.033 |
| 125 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
-0.294 |
0.061 |
| 126 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.294 |
-0.527 |
| 127 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
-0.290 |
-0.533 |
| 128 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.289 |
-0.330 |
| 129 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.289 |
-0.373 |
| 130 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.281 |
-0.120 |
| 131 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.277 |
-0.019 |
| 132 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.275 |
-0.288 |
| 133 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.258 |
-0.599 |
| 134 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
-0.257 |
-0.266 |
| 135 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.256 |
-0.942 |
| 136 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
-0.255 |
-0.081 |
| 137 |
253305_at |
AT4G33666
|
unknown |
-0.244 |
-0.124 |
| 138 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.243 |
-0.413 |
| 139 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.243 |
0.021 |
| 140 |
258421_at |
AT3G16690
|
AtSWEET16, SWEET16 |
-0.242 |
-0.480 |
| 141 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
-0.242 |
-0.264 |
| 142 |
259373_at |
AT1G69160
|
unknown |
-0.240 |
-0.081 |
| 143 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.238 |
-0.060 |
| 144 |
256021_at |
AT1G58270
|
ZW9 |
-0.237 |
0.282 |
| 145 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.237 |
-0.323 |
| 146 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.236 |
-0.096 |
| 147 |
246487_at |
AT5G16030
|
unknown |
-0.233 |
-0.462 |
| 148 |
250223_at |
AT5G14070
|
ROXY2 |
-0.228 |
-0.241 |
| 149 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.228 |
-0.224 |
| 150 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.227 |
0.029 |
| 151 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.227 |
-0.385 |
| 152 |
254787_at |
AT4G12690
|
unknown |
-0.225 |
-0.313 |
| 153 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.225 |
-0.564 |
| 154 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.219 |
-0.025 |
| 155 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
-0.218 |
0.126 |
| 156 |
246783_at |
AT5G27360
|
SFP2 |
-0.214 |
-0.463 |
| 157 |
261451_at |
AT1G21060
|
unknown |
-0.214 |
-0.066 |
| 158 |
252215_at |
AT3G50240
|
KICP-02 |
-0.207 |
-0.414 |
| 159 |
245016_at |
ATCG00500
|
ACCD, acetyl-CoA carboxylase carboxyl transferase subunit beta |
-0.207 |
-0.062 |
| 160 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.206 |
0.293 |
| 161 |
261983_at |
AT1G33770
|
[Protein kinase superfamily protein] |
-0.205 |
-0.677 |
| 162 |
251306_at |
AT3G61260
|
[Remorin family protein] |
-0.204 |
-0.129 |
| 163 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.204 |
-0.216 |
| 164 |
258468_at |
AT3G06070
|
unknown |
-0.200 |
-1.076 |
| 165 |
258196_at |
AT3G13980
|
unknown |
-0.198 |
-0.168 |
| 166 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.197 |
-0.486 |
| 167 |
257317_at |
ATMG01060
|
ORF107G |
-0.195 |
0.007 |
| 168 |
253408_at |
AT4G32950
|
[Protein phosphatase 2C family protein] |
-0.194 |
-0.027 |
| 169 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.194 |
-0.182 |
| 170 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.193 |
-0.108 |
| 171 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
-0.191 |
-0.060 |
| 172 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.190 |
-0.300 |
| 173 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
-0.187 |
0.226 |
| 174 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.186 |
-0.665 |
| 175 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
-0.186 |
-0.022 |
| 176 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.186 |
-0.502 |
| 177 |
259182_at |
AT3G01750
|
[Ankyrin repeat family protein] |
-0.183 |
-0.118 |
| 178 |
267209_at |
AT2G30930
|
unknown |
-0.183 |
-0.450 |
| 179 |
258225_at |
AT3G15630
|
unknown |
-0.182 |
-0.345 |
| 180 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-0.180 |
-0.123 |
| 181 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.179 |
-1.304 |
| 182 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-0.179 |
-0.199 |
| 183 |
266917_at |
AT2G45830
|
DTA2, downstream target of AGL15 2 |
-0.178 |
-0.082 |
| 184 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.176 |
-0.428 |
| 185 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.175 |
-0.114 |
| 186 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.175 |
-0.591 |
| 187 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.174 |
-0.515 |
| 188 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.172 |
-0.298 |
| 189 |
250189_at |
AT5G14410
|
unknown |
-0.172 |
0.010 |
| 190 |
256010_at |
AT1G19220
|
ARF11, AUXIN RESPONSE FACTOR11, ARF19, auxin response factor 19, IAA22, indole-3-acetic acid inducible 22 |
-0.172 |
-0.070 |
| 191 |
250697_at |
AT5G06800
|
[myb-like HTH transcriptional regulator family protein] |
-0.171 |
-0.108 |
| 192 |
245138_at |
AT2G45190
|
AFO, ABNORMAL FLORAL ORGANS, FIL, FILAMENTOUS FLOWER, YAB1, YABBY1 |
-0.169 |
-0.258 |
| 193 |
264822_at |
AT1G03457
|
AtBRN2, BRN2, Bruno-like 2 |
-0.167 |
-0.455 |
| 194 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.167 |
-0.681 |
| 195 |
266715_at |
AT2G46780
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.167 |
-0.592 |
| 196 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.165 |
-0.725 |
| 197 |
248204_at |
AT5G54280
|
ATM2, myosin 2, ATM4, ARABIDOPSIS THALIANA MYOSIN 4, ATMYOS1, ARABIDOPSIS THALIANA MYOSIN 1 |
-0.164 |
0.228 |
| 198 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.164 |
-0.204 |
| 199 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.164 |
-0.525 |
| 200 |
246926_at |
AT5G25240
|
unknown |
-0.163 |
-0.042 |
| 201 |
259541_at |
AT1G20650
|
ASG5, ALTERED SEED GERMINATION 5 |
-0.162 |
-0.029 |