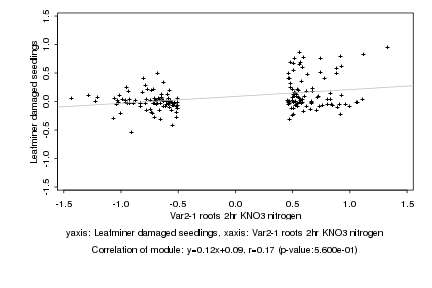
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Var2-1 roots 2hr KNO3 nitrogen |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
1.324 |
0.955 |
| 2 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
1.114 |
0.836 |
| 3 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
1.105 |
0.039 |
| 4 |
248122_at |
AT5G54700
|
[Ankyrin repeat family protein] |
1.062 |
-0.013 |
| 5 |
262421_at |
AT1G50290
|
unknown |
1.055 |
-0.009 |
| 6 |
244973_at |
ATCG00690
|
PSBT, photosystem II reaction center protein T, PSBTC |
0.991 |
-0.074 |
| 7 |
244994_at |
ATCG01010
|
NDHF |
0.956 |
-0.052 |
| 8 |
246406_at |
AT1G57650
|
[ATP binding] |
0.922 |
0.114 |
| 9 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.919 |
0.627 |
| 10 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.912 |
0.797 |
| 11 |
257168_at |
AT3G24430
|
HCF101, HIGH-CHLOROPHYLL-FLUORESCENCE 101 |
0.910 |
-0.213 |
| 12 |
263769_at |
AT2G06390
|
transposable element gene |
0.902 |
-0.045 |
| 13 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.889 |
-0.094 |
| 14 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.875 |
0.597 |
| 15 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.875 |
0.500 |
| 16 |
244965_at |
ATCG00590
|
ORF31 |
0.842 |
-0.056 |
| 17 |
244933_at |
ATCG01070
|
NDHE |
0.839 |
-0.051 |
| 18 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
0.829 |
0.052 |
| 19 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.823 |
0.156 |
| 20 |
245135_at |
AT2G45230
|
[non-LTR retrotransposon family (LINE), has a 3.9e-46 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
0.803 |
-0.041 |
| 21 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
0.796 |
0.048 |
| 22 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.776 |
0.414 |
| 23 |
260092_at |
AT1G73280
|
scpl3, serine carboxypeptidase-like 3 |
0.756 |
-0.061 |
| 24 |
256442_at |
AT3G10930
|
unknown |
0.735 |
0.757 |
| 25 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.735 |
0.518 |
| 26 |
244934_at |
ATCG01080
|
NDHG |
0.730 |
-0.080 |
| 27 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.721 |
0.091 |
| 28 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.709 |
0.073 |
| 29 |
265483_at |
AT2G15790
|
CYP40, CYCLOPHILIN 40, SQN, SQUINT |
0.707 |
-0.156 |
| 30 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.673 |
0.245 |
| 31 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.666 |
0.184 |
| 32 |
257354_x_at |
AT2G23480
|
unknown |
0.658 |
0.005 |
| 33 |
250135_at |
AT5G15360
|
unknown |
0.656 |
-0.011 |
| 34 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
0.656 |
-0.020 |
| 35 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
0.650 |
-0.132 |
| 36 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.624 |
0.476 |
| 37 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.613 |
-0.085 |
| 38 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.613 |
0.190 |
| 39 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.595 |
0.090 |
| 40 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.591 |
0.773 |
| 41 |
244997_at |
ATCG00170
|
RPOC2 |
0.587 |
-0.174 |
| 42 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
0.584 |
0.053 |
| 43 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
0.581 |
0.611 |
| 44 |
254596_at |
AT4G18975
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.580 |
-0.016 |
| 45 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.569 |
0.361 |
| 46 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
0.569 |
-0.026 |
| 47 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.564 |
0.685 |
| 48 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
0.562 |
0.033 |
| 49 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.559 |
0.058 |
| 50 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.559 |
0.864 |
| 51 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.557 |
0.667 |
| 52 |
254976_at |
AT4G10510
|
[Subtilase family protein] |
0.553 |
-0.004 |
| 53 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.550 |
0.210 |
| 54 |
244903_at |
ATMG00660
|
ORF149 |
0.550 |
0.075 |
| 55 |
260975_at |
AT1G53430
|
[Leucine-rich repeat transmembrane protein kinase] |
0.540 |
0.212 |
| 56 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.538 |
-0.025 |
| 57 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
0.535 |
-0.070 |
| 58 |
244921_s_at |
ATMG01000
|
ORF114 |
0.529 |
0.135 |
| 59 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.525 |
0.097 |
| 60 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.525 |
-0.049 |
| 61 |
244932_at |
ATCG01060
|
PSAC |
0.524 |
-0.063 |
| 62 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
0.523 |
0.143 |
| 63 |
254566_at |
AT4G19240
|
unknown |
0.519 |
0.009 |
| 64 |
245082_at |
AT2G23270
|
unknown |
0.514 |
0.172 |
| 65 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.514 |
0.767 |
| 66 |
252036_at |
AT3G52070
|
unknown |
0.512 |
0.095 |
| 67 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.506 |
0.557 |
| 68 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.504 |
0.677 |
| 69 |
245008_at |
ATCG00360
|
YCF3 |
0.502 |
-0.213 |
| 70 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
0.502 |
0.140 |
| 71 |
244966_at |
ATCG00600
|
PETG |
0.501 |
-0.107 |
| 72 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
0.499 |
-0.017 |
| 73 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.496 |
0.214 |
| 74 |
253796_at |
AT4G28460
|
unknown |
0.495 |
0.082 |
| 75 |
265387_at |
AT2G20670
|
unknown |
0.495 |
-0.242 |
| 76 |
262004_at |
AT1G64480
|
CBL8, calcineurin B-like protein 8 |
0.490 |
0.019 |
| 77 |
248539_at |
AT5G50130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.487 |
-0.117 |
| 78 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.481 |
0.701 |
| 79 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.479 |
0.251 |
| 80 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
0.474 |
0.330 |
| 81 |
245737_at |
AT1G44160
|
[HSP40/DnaJ peptide-binding protein] |
0.471 |
0.007 |
| 82 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
0.471 |
0.413 |
| 83 |
260073_at |
AT1G73660
|
SIS8, SUGAR INSENSITIVE 8 |
0.466 |
-0.315 |
| 84 |
246316_at |
AT3G56890
|
[F-box associated ubiquitination effector family protein] |
0.465 |
-0.029 |
| 85 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.458 |
0.011 |
| 86 |
247807_at |
AT5G58360
|
ATOFP3, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 3, OFP3, ovate family protein 3 |
0.458 |
-0.048 |
| 87 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.458 |
0.496 |
| 88 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
0.456 |
0.406 |
| 89 |
261558_at |
AT1G01770
|
unknown |
0.455 |
-0.012 |
| 90 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
0.454 |
0.026 |
| 91 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
-1.439 |
0.023 |
| 92 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-1.290 |
0.111 |
| 93 |
259260_at |
AT3G11370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.231 |
0.008 |
| 94 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
-1.212 |
0.077 |
| 95 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
-1.071 |
-0.287 |
| 96 |
267267_at |
AT2G02580
|
CYP71B9, cytochrome P450, family 71, subfamily B, polypeptide 9 |
-1.059 |
0.061 |
| 97 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-1.047 |
-0.041 |
| 98 |
267572_at |
AT2G30660
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
-1.035 |
0.019 |
| 99 |
248183_at |
AT5G54040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.024 |
-0.009 |
| 100 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-1.015 |
0.108 |
| 101 |
246393_at |
AT1G58150
|
unknown |
-1.010 |
-0.200 |
| 102 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.993 |
0.050 |
| 103 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
-0.965 |
0.023 |
| 104 |
266307_at |
AT2G27000
|
CYP705A8, cytochrome P450, family 705, subfamily A, polypeptide 8 |
-0.965 |
-0.015 |
| 105 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
-0.953 |
0.262 |
| 106 |
261091_at |
AT1G07550
|
[Leucine-rich repeat protein kinase family protein] |
-0.947 |
-0.018 |
| 107 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
-0.936 |
0.186 |
| 108 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
-0.932 |
-0.019 |
| 109 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
-0.930 |
0.037 |
| 110 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.914 |
-0.543 |
| 111 |
246374_at |
AT1G51840
|
[protein kinase-related] |
-0.894 |
-0.021 |
| 112 |
248212_at |
AT5G54020
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.879 |
0.019 |
| 113 |
252458_at |
AT3G47210
|
unknown |
-0.836 |
-0.031 |
| 114 |
257617_at |
AT3G26550
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.832 |
-0.077 |
| 115 |
252196_at |
AT3G50200
|
unknown |
-0.831 |
-0.024 |
| 116 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
-0.816 |
0.162 |
| 117 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
-0.809 |
0.417 |
| 118 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
-0.791 |
-0.022 |
| 119 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.786 |
0.291 |
| 120 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.784 |
-0.143 |
| 121 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.782 |
0.043 |
| 122 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.775 |
0.224 |
| 123 |
254979_at |
AT4G10550
|
[Subtilase family protein] |
-0.747 |
-0.125 |
| 124 |
265124_at |
AT1G55430
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.743 |
0.020 |
| 125 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
-0.734 |
0.200 |
| 126 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.734 |
-0.181 |
| 127 |
265870_at |
AT2G01660
|
PDLP6, plasmodesmata-located protein 6 |
-0.726 |
-0.203 |
| 128 |
252490_at |
AT3G46720
|
[UDP-Glycosyltransferase superfamily protein] |
-0.723 |
-0.019 |
| 129 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
-0.719 |
-0.030 |
| 130 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
-0.718 |
0.226 |
| 131 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.716 |
-0.270 |
| 132 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
-0.714 |
0.063 |
| 133 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.708 |
-0.025 |
| 134 |
245226_at |
AT3G29970
|
[B12D protein] |
-0.704 |
0.029 |
| 135 |
255547_at |
AT4G01920
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.696 |
-0.040 |
| 136 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.693 |
0.029 |
| 137 |
261879_at |
AT1G50520
|
CYP705A27, cytochrome P450, family 705, subfamily A, polypeptide 27 |
-0.688 |
-0.022 |
| 138 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
-0.685 |
0.509 |
| 139 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.679 |
0.062 |
| 140 |
265559_at |
AT2G05530
|
[Glycine-rich protein family] |
-0.672 |
0.043 |
| 141 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
-0.666 |
-0.156 |
| 142 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
-0.662 |
-0.313 |
| 143 |
266176_at |
AT2G02250
|
AtPP2-B2, phloem protein 2-B2, PP2-B2, phloem protein 2-B2 |
-0.661 |
-0.020 |
| 144 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.653 |
0.137 |
| 145 |
256684_at |
AT3G32040
|
[Terpenoid synthases superfamily protein] |
-0.648 |
0.083 |
| 146 |
253372_at |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
-0.641 |
0.022 |
| 147 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.635 |
0.345 |
| 148 |
254510_at |
AT4G20210
|
TPS08, terpene synthase 8 |
-0.633 |
0.019 |
| 149 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.631 |
0.017 |
| 150 |
255059_at |
AT4G09420
|
[Disease resistance protein (TIR-NBS class)] |
-0.629 |
-0.071 |
| 151 |
245270_at |
AT4G14960
|
TUA6, Tubulin alpha-6 |
-0.616 |
-0.079 |
| 152 |
265290_at |
AT2G22590
|
[UDP-Glycosyltransferase superfamily protein] |
-0.609 |
-0.050 |
| 153 |
254743_at |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
-0.607 |
0.007 |
| 154 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
-0.594 |
0.129 |
| 155 |
263145_at |
AT1G54090
|
ATEXO70D2, exocyst subunit exo70 family protein D2, EXO70D2, exocyst subunit exo70 family protein D2 |
-0.589 |
0.029 |
| 156 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
-0.588 |
0.069 |
| 157 |
248077_at |
AT5G55770
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.587 |
-0.024 |
| 158 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
-0.586 |
-0.052 |
| 159 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.580 |
0.198 |
| 160 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.579 |
-0.100 |
| 161 |
246992_at |
AT5G67430
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.578 |
-0.005 |
| 162 |
245553_at |
AT4G15370
|
BARS1, baruol synthase 1, PEN2, PENTACYCLIC TRITERPENE SYNTHASE 2 |
-0.575 |
0.027 |
| 163 |
250771_at |
AT5G05400
|
[LRR and NB-ARC domains-containing disease resistance protein] |
-0.570 |
0.016 |
| 164 |
251044_at |
AT5G02350
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.566 |
-0.010 |
| 165 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.560 |
-0.156 |
| 166 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.558 |
-0.027 |
| 167 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.555 |
-0.411 |
| 168 |
258769_at |
AT3G10870
|
ATMES17, ARABIDOPSIS THALIANA METHYL ESTERASE 17, MES17, methyl esterase 17 |
-0.552 |
-0.055 |
| 169 |
245612_at |
AT4G14440
|
ATECI3, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, ECI3, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, HCD1, 3-hydroxyacyl-CoA dehydratase 1 |
-0.542 |
-0.065 |
| 170 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.539 |
-0.017 |
| 171 |
264101_at |
AT1G79000
|
ATHAC1, ARABIDOPSIS HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY 1, ATHPCAT2, ARABIDOPSIS THALIANA P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 2, HAC1, histone acetyltransferase of the CBP family 1, PCAT2, P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 2 |
-0.534 |
-0.032 |
| 172 |
265151_at |
AT1G51340
|
[MATE efflux family protein] |
-0.519 |
-0.266 |
| 173 |
257112_at |
AT3G20120
|
CYP705A21, cytochrome P450, family 705, subfamily A, polypeptide 21 |
-0.518 |
-0.076 |
| 174 |
249245_at |
AT5G42280
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.516 |
-0.191 |
| 175 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.515 |
-0.113 |
| 176 |
254320_at |
AT4G22580
|
[Exostosin family protein] |
-0.514 |
-0.064 |
| 177 |
254902_at |
AT4G11550
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.514 |
-0.107 |
| 178 |
256162_at |
AT1G55390
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.512 |
-0.017 |
| 179 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.511 |
0.068 |