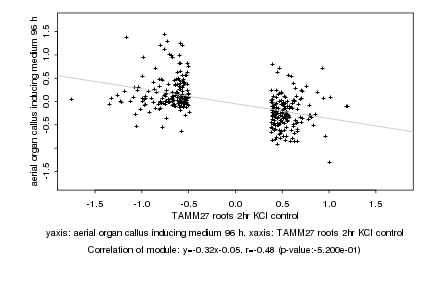
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
TAMM27 roots 2hr KCl control |
Log2 signal ratio
aerial organ callus inducing medium 96 h |
| 1 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
1.194 |
-0.094 |
| 2 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
1.181 |
-0.099 |
| 3 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
1.013 |
0.094 |
| 4 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.001 |
-1.287 |
| 5 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
0.961 |
-0.741 |
| 6 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.932 |
0.065 |
| 7 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.929 |
0.728 |
| 8 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.874 |
0.201 |
| 9 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.849 |
-0.269 |
| 10 |
245354_at |
AT4G17600
|
LIL3:1 |
0.825 |
-0.496 |
| 11 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.804 |
-0.338 |
| 12 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
0.792 |
-0.284 |
| 13 |
248122_at |
AT5G54700
|
[Ankyrin repeat family protein] |
0.789 |
-0.075 |
| 14 |
265998_at |
AT2G24270
|
ALDH11A3, aldehyde dehydrogenase 11A3 |
0.786 |
-0.285 |
| 15 |
254727_at |
AT4G13670
|
PTAC5, plastid transcriptionally active 5 |
0.774 |
-0.379 |
| 16 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.756 |
0.336 |
| 17 |
265481_at |
AT2G15960
|
unknown |
0.714 |
0.236 |
| 18 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
0.706 |
-0.354 |
| 19 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
0.705 |
-0.445 |
| 20 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.700 |
0.251 |
| 21 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.695 |
0.078 |
| 22 |
259572_at |
AT1G20400
|
unknown |
0.688 |
0.058 |
| 23 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
0.686 |
-0.328 |
| 24 |
261581_at |
AT1G01140
|
CIPK9, CBL-interacting protein kinase 9, PKS6, PROTEIN KINASE 6, SnRK3.12, SNF1-RELATED PROTEIN KINASE 3.12 |
0.665 |
-0.047 |
| 25 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
0.662 |
-0.593 |
| 26 |
248177_at |
AT5G54630
|
[zinc finger protein-related] |
0.662 |
-0.448 |
| 27 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
0.658 |
-0.857 |
| 28 |
246888_at |
AT5G26270
|
unknown |
0.658 |
-0.171 |
| 29 |
257168_at |
AT3G24430
|
HCF101, HIGH-CHLOROPHYLL-FLUORESCENCE 101 |
0.653 |
-0.640 |
| 30 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.650 |
0.126 |
| 31 |
244997_at |
ATCG00170
|
RPOC2 |
0.642 |
-0.456 |
| 32 |
265483_at |
AT2G15790
|
CYP40, CYCLOPHILIN 40, SQN, SQUINT |
0.641 |
0.290 |
| 33 |
252663_at |
AT3G44070
|
[Glycosyl hydrolase family 35 protein] |
0.633 |
-0.389 |
| 34 |
254565_at |
AT4G19130
|
[Replication factor-A protein 1-related] |
0.626 |
0.032 |
| 35 |
252694_at |
AT3G43630
|
[Vacuolar iron transporter (VIT) family protein] |
0.624 |
0.014 |
| 36 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.622 |
-0.842 |
| 37 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
0.621 |
-0.679 |
| 38 |
253689_at |
AT4G29770
|
unknown |
0.618 |
0.033 |
| 39 |
247320_at |
AT5G64040
|
PSAN |
0.615 |
-0.518 |
| 40 |
244901_at |
ATMG00640
|
ORF25 |
0.611 |
0.402 |
| 41 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.610 |
-0.472 |
| 42 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
0.609 |
-0.021 |
| 43 |
244903_at |
ATMG00660
|
ORF149 |
0.607 |
-0.028 |
| 44 |
249524_at |
AT5G38520
|
[alpha/beta-Hydrolases superfamily protein] |
0.602 |
-0.787 |
| 45 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.599 |
-0.140 |
| 46 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.598 |
0.545 |
| 47 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.593 |
-0.127 |
| 48 |
250257_at |
AT5G13770
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.591 |
-0.601 |
| 49 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
0.578 |
-0.850 |
| 50 |
267526_at |
AT2G30570
|
PSBW, photosystem II reaction center W |
0.574 |
-0.339 |
| 51 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
0.571 |
-0.149 |
| 52 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
0.568 |
-0.097 |
| 53 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
0.566 |
-0.301 |
| 54 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
0.565 |
-0.285 |
| 55 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.558 |
0.575 |
| 56 |
245701_at |
AT5G04140
|
FD-GOGAT, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE, GLS1, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1, GLU1, glutamate synthase 1, GLUS |
0.556 |
-0.404 |
| 57 |
262536_at |
AT1G17100
|
AtHBP1, HBP1, haem-binding protein 1 |
0.554 |
-0.413 |
| 58 |
267376_at |
AT2G26330
|
ER, ERECTA, QRP1, QUANTITATIVE RESISTANCE TO PLECTOSPHAERELLA 1 |
0.551 |
-0.278 |
| 59 |
249887_at |
AT5G22310
|
unknown |
0.548 |
0.021 |
| 60 |
252155_at |
AT3G50920
|
LPPepsilon1, lipid phosphate phosphatase epsilon 1 |
0.547 |
-0.098 |
| 61 |
245213_at |
AT1G44575
|
CP22, NPQ4, NONPHOTOCHEMICAL QUENCHING 4, PSBS, PHOTOSYSTEM II SUBUNIT S |
0.544 |
-0.550 |
| 62 |
259707_at |
AT1G77490
|
TAPX, thylakoidal ascorbate peroxidase |
0.539 |
-0.740 |
| 63 |
246417_at |
AT5G16990
|
[Zinc-binding dehydrogenase family protein] |
0.538 |
-0.201 |
| 64 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
0.536 |
-0.353 |
| 65 |
267299_at |
AT2G30150
|
[UDP-Glycosyltransferase superfamily protein] |
0.535 |
-0.262 |
| 66 |
261544_at |
AT1G63540
|
[hydroxyproline-rich glycoprotein family protein] |
0.534 |
-0.143 |
| 67 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
0.533 |
-0.040 |
| 68 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.533 |
0.088 |
| 69 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
0.533 |
-0.336 |
| 70 |
252409_at |
AT3G47650
|
[DnaJ/Hsp40 cysteine-rich domain superfamily protein] |
0.530 |
-0.352 |
| 71 |
253237_at |
AT4G34240
|
ALDH3, aldehyde dehydrogenase 3, ALDH3I1, aldehyde dehydrogenase 3I1 |
0.526 |
-0.277 |
| 72 |
260696_at |
AT1G32520
|
unknown |
0.525 |
-0.822 |
| 73 |
265287_at |
AT2G20260
|
PSAE-2, photosystem I subunit E-2 |
0.524 |
-0.332 |
| 74 |
261861_at |
AT1G50450
|
[Saccharopine dehydrogenase ] |
0.520 |
-0.453 |
| 75 |
249715_at |
AT5G35760
|
[Beta-galactosidase related protein] |
0.520 |
-0.087 |
| 76 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
0.519 |
0.126 |
| 77 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.519 |
-0.315 |
| 78 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
0.516 |
-0.612 |
| 79 |
259499_at |
AT1G15730
|
[Cobalamin biosynthesis CobW-like protein] |
0.514 |
-0.341 |
| 80 |
253860_at |
AT4G27700
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.509 |
-0.731 |
| 81 |
255760_at |
AT1G16780
|
AtVHP2;2, VHP2;2 |
0.506 |
-0.163 |
| 82 |
254537_at |
AT4G19730
|
[Glycosyl hydrolase superfamily protein] |
0.505 |
-0.043 |
| 83 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.505 |
-0.003 |
| 84 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.505 |
-0.364 |
| 85 |
259363_at |
AT1G13270
|
MAP1B, METHIONINE AMINOPEPTIDASE 1B, MAP1C, methionine aminopeptidase 1B |
0.501 |
-0.252 |
| 86 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
0.497 |
-0.177 |
| 87 |
245026_at |
ATCG00140
|
ATPH |
0.496 |
-0.313 |
| 88 |
245025_at |
ATCG00130
|
ATPF |
0.494 |
-0.434 |
| 89 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
0.493 |
-0.055 |
| 90 |
257447_at |
AT2G04230
|
[FBD, F-box and Leucine Rich Repeat domains containing protein] |
0.493 |
-0.230 |
| 91 |
261540_at |
AT1G63610
|
unknown |
0.492 |
-0.391 |
| 92 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.490 |
-0.643 |
| 93 |
253796_at |
AT4G28460
|
unknown |
0.490 |
0.033 |
| 94 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
0.487 |
-0.054 |
| 95 |
264356_at |
AT1G03190
|
ATXPD, ARABIDOPSIS THALIANA XERODERMA PIGMENTOSUM GROUP D, UVH6, ULTRAVIOLET HYPERSENSITIVE 6 |
0.484 |
0.188 |
| 96 |
259730_at |
AT1G77660
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.483 |
0.196 |
| 97 |
253814_at |
AT4G28290
|
unknown |
0.480 |
-0.282 |
| 98 |
256309_at |
AT1G30380
|
PSAK, photosystem I subunit K |
0.480 |
-0.429 |
| 99 |
245008_at |
ATCG00360
|
YCF3 |
0.478 |
-0.396 |
| 100 |
259193_at |
AT3G01480
|
ATCYP38, ARABIDOPSIS CYCLOPHILIN 38, CYP38, cyclophilin 38 |
0.476 |
-0.669 |
| 101 |
250075_at |
AT5G17670
|
[alpha/beta-Hydrolases superfamily protein] |
0.474 |
-0.789 |
| 102 |
247319_at |
AT5G64050
|
ATERS, ERS, glutamate tRNA synthetase, OVA3, OVULE ABORTION 3 |
0.473 |
-0.246 |
| 103 |
250073_at |
AT5G17170
|
ENH1, enhancer of sos3-1 |
0.469 |
-0.690 |
| 104 |
254623_at |
AT4G18480
|
CH-42, CHLORINA 42, CH42, CHLORINA 42, CHL11, CHLI-1, CHLI1 |
0.467 |
-0.478 |
| 105 |
262821_at |
AT1G11800
|
[endonuclease/exonuclease/phosphatase family protein] |
0.466 |
0.008 |
| 106 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.465 |
0.722 |
| 107 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.464 |
0.185 |
| 108 |
248287_at |
AT5G52970
|
[thylakoid lumen 15.0 kDa protein] |
0.460 |
-0.607 |
| 109 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
0.459 |
-0.500 |
| 110 |
250135_at |
AT5G15360
|
unknown |
0.457 |
-0.337 |
| 111 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
0.455 |
-0.139 |
| 112 |
255733_at |
AT1G25400
|
unknown |
0.454 |
0.171 |
| 113 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
0.449 |
-0.904 |
| 114 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
0.447 |
-0.353 |
| 115 |
258804_at |
AT3G04760
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.443 |
-0.463 |
| 116 |
257078_at |
AT3G15220
|
[Protein kinase superfamily protein] |
0.441 |
-0.248 |
| 117 |
267172_at |
AT2G37660
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.441 |
-0.507 |
| 118 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
0.439 |
0.628 |
| 119 |
254182_at |
AT4G23950
|
[Galactose-binding protein] |
0.438 |
-0.110 |
| 120 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.438 |
-0.224 |
| 121 |
249321_at |
AT5G40920
|
[pseudogene] |
0.438 |
-0.204 |
| 122 |
252552_at |
AT3G45900
|
[Ribonuclease P protein subunit P38-related] |
0.435 |
-0.209 |
| 123 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.434 |
0.242 |
| 124 |
253532_at |
AT4G31570
|
unknown |
0.434 |
0.103 |
| 125 |
260236_at |
AT1G74470
|
[Pyridine nucleotide-disulphide oxidoreductase family protein] |
0.431 |
-0.258 |
| 126 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
0.430 |
-0.751 |
| 127 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.430 |
-0.038 |
| 128 |
246308_at |
AT3G51820
|
ATG4, CHLG, G4, PDE325, PIGMENT DEFECTIVE 325 |
0.427 |
-0.581 |
| 129 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.427 |
-0.332 |
| 130 |
245004_at |
ATCG00300
|
YCF9 |
0.426 |
-0.207 |
| 131 |
253052_at |
AT4G37310
|
CYP81H1, cytochrome P450, family 81, subfamily H, polypeptide 1 |
0.422 |
-0.043 |
| 132 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
0.422 |
-0.509 |
| 133 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
0.421 |
-0.801 |
| 134 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
0.416 |
-0.257 |
| 135 |
252516_at |
AT3G46100
|
ATHRS1, Histidyl-tRNA synthetase 1, HRS1, Histidyl-tRNA synthetase 1 |
0.416 |
-0.240 |
| 136 |
254970_at |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
0.415 |
-0.279 |
| 137 |
258285_at |
AT3G16140
|
PSAH-1, photosystem I subunit H-1 |
0.414 |
-0.395 |
| 138 |
261119_at |
AT1G75350
|
emb2184, embryo defective 2184 |
0.413 |
-0.320 |
| 139 |
257932_at |
AT3G17040
|
HCF107, high chlorophyll fluorescent 107 |
0.413 |
-0.606 |
| 140 |
249066_at |
AT5G43920
|
[transducin family protein / WD-40 repeat family protein] |
0.412 |
-0.126 |
| 141 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
0.411 |
-0.467 |
| 142 |
256442_at |
AT3G10930
|
unknown |
0.408 |
-0.109 |
| 143 |
244921_s_at |
ATMG01000
|
ORF114 |
0.408 |
-0.107 |
| 144 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.405 |
0.125 |
| 145 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
0.401 |
-0.147 |
| 146 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.400 |
-0.237 |
| 147 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
0.399 |
-0.483 |
| 148 |
261075_at |
AT1G07280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.399 |
-0.073 |
| 149 |
264920_at |
AT1G60550
|
DHNS, ECHID, enoyl-CoA hydratase/isomerase D |
0.396 |
-0.476 |
| 150 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
0.394 |
-0.571 |
| 151 |
262411_at |
AT1G34640
|
[peptidases] |
0.394 |
-0.217 |
| 152 |
245006_at |
ATCG00340
|
PSAB |
0.394 |
-0.105 |
| 153 |
262875_at |
AT1G64970
|
G-TMT, gamma-tocopherol methyltransferase, TMT1, VTE4, VITAMIN E DEFICIENT 4 |
0.393 |
-0.384 |
| 154 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
0.391 |
-0.559 |
| 155 |
248634_at |
AT5G49030
|
OVA2, ovule abortion 2 |
0.391 |
-0.297 |
| 156 |
253279_at |
AT4G34030
|
MCCB, 3-methylcrotonyl-CoA carboxylase |
0.390 |
0.062 |
| 157 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.389 |
-0.009 |
| 158 |
262558_at |
AT1G31335
|
unknown |
0.388 |
-0.175 |
| 159 |
266756_at |
AT2G46950
|
CYP709B2, cytochrome P450, family 709, subfamily B, polypeptide 2 |
0.388 |
0.803 |
| 160 |
256652_at |
AT3G18850
|
LPAT5, lysophosphatidyl acyltransferase 5 |
0.388 |
-0.103 |
| 161 |
250190_at |
AT5G14320
|
EMB3137, EMBRYO DEFECTIVE 3137 |
0.386 |
-0.306 |
| 162 |
251784_at |
AT3G55330
|
PPL1, PsbP-like protein 1 |
0.386 |
-0.834 |
| 163 |
254566_at |
AT4G19240
|
unknown |
0.385 |
0.043 |
| 164 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
0.385 |
-0.433 |
| 165 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.385 |
-0.658 |
| 166 |
246817_at |
AT5G27240
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.382 |
-0.017 |
| 167 |
246847_at |
AT5G26820
|
ATIREG3, iron-regulated protein 3, IREG3, IRON REGULATED 3, IREG3, iron-regulated protein 3, MAR1, MULTIPLE ANTIBIOTIC RESISTANCE 1, RTS3 |
0.380 |
-0.549 |
| 168 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.380 |
0.249 |
| 169 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
-1.759 |
0.046 |
| 170 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
-1.348 |
-0.062 |
| 171 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-1.329 |
0.066 |
| 172 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-1.268 |
0.144 |
| 173 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
-1.231 |
0.015 |
| 174 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
-1.219 |
-0.018 |
| 175 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
-1.186 |
0.215 |
| 176 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-1.174 |
1.389 |
| 177 |
267267_at |
AT2G02580
|
CYP71B9, cytochrome P450, family 71, subfamily B, polypeptide 9 |
-1.123 |
0.004 |
| 178 |
248183_at |
AT5G54040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.095 |
0.105 |
| 179 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-1.082 |
0.319 |
| 180 |
245267_at |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.069 |
-0.257 |
| 181 |
246393_at |
AT1G58150
|
unknown |
-1.067 |
-0.530 |
| 182 |
266307_at |
AT2G27000
|
CYP705A8, cytochrome P450, family 705, subfamily A, polypeptide 8 |
-1.048 |
0.247 |
| 183 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.040 |
0.308 |
| 184 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-1.018 |
-0.160 |
| 185 |
245554_at |
AT4G15380
|
CYP705A4, cytochrome P450, family 705, subfamily A, polypeptide 4 |
-1.000 |
0.016 |
| 186 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
-0.999 |
0.082 |
| 187 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
-0.999 |
0.550 |
| 188 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.985 |
0.962 |
| 189 |
246374_at |
AT1G51840
|
[protein kinase-related] |
-0.982 |
0.058 |
| 190 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
-0.972 |
-0.076 |
| 191 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-0.969 |
0.081 |
| 192 |
267572_at |
AT2G30660
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
-0.968 |
-0.044 |
| 193 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
-0.965 |
0.115 |
| 194 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
-0.934 |
0.218 |
| 195 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.927 |
-0.220 |
| 196 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.903 |
0.085 |
| 197 |
252232_at |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
-0.883 |
-0.010 |
| 198 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.877 |
0.412 |
| 199 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.871 |
0.273 |
| 200 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.861 |
-0.143 |
| 201 |
252490_at |
AT3G46720
|
[UDP-Glycosyltransferase superfamily protein] |
-0.860 |
0.720 |
| 202 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.852 |
0.198 |
| 203 |
255811_at |
AT4G10250
|
ATHSP22.0 |
-0.825 |
-0.052 |
| 204 |
248212_at |
AT5G54020
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.821 |
0.333 |
| 205 |
266827_at |
AT2G22920
|
SCPL12, serine carboxypeptidase-like 12 |
-0.817 |
-0.157 |
| 206 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.814 |
0.484 |
| 207 |
261091_at |
AT1G07550
|
[Leucine-rich repeat protein kinase family protein] |
-0.807 |
-0.020 |
| 208 |
249617_at |
AT5G37450
|
[Leucine-rich repeat protein kinase family protein] |
-0.807 |
-0.139 |
| 209 |
263572_at |
AT2G17060
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.805 |
-0.017 |
| 210 |
245674_at |
AT1G56680
|
[Chitinase family protein] |
-0.804 |
1.212 |
| 211 |
254979_at |
AT4G10550
|
[Subtilase family protein] |
-0.802 |
0.001 |
| 212 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
-0.798 |
-0.024 |
| 213 |
252882_at |
AT4G39675
|
unknown |
-0.790 |
0.491 |
| 214 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
-0.788 |
0.470 |
| 215 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.788 |
0.080 |
| 216 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.781 |
-0.542 |
| 217 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
-0.771 |
-0.026 |
| 218 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
-0.767 |
1.455 |
| 219 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.760 |
1.126 |
| 220 |
257085_at |
AT3G20630
|
ATUBP14, PER1, phosphate deficiency root hair defective1, TTN6, TITAN6, UBP14, ubiquitin-specific protease 14 |
-0.759 |
0.142 |
| 221 |
245556_at |
AT4G15400
|
ABS1, ABNORMAL SHOOT 1, BIA1, BRASSINOSTEROID INACTIVATOR1 |
-0.757 |
0.020 |
| 222 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
-0.757 |
0.124 |
| 223 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
-0.753 |
-0.047 |
| 224 |
255547_at |
AT4G01920
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.751 |
0.168 |
| 225 |
263023_at |
AT1G23960
|
unknown |
-0.747 |
0.007 |
| 226 |
256563_at |
AT3G29780
|
RALFL27, ralf-like 27 |
-0.746 |
0.250 |
| 227 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
-0.743 |
-0.348 |
| 228 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
-0.742 |
0.006 |
| 229 |
254510_at |
AT4G20210
|
TPS08, terpene synthase 8 |
-0.740 |
0.175 |
| 230 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
-0.734 |
1.300 |
| 231 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.723 |
0.368 |
| 232 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.722 |
0.061 |
| 233 |
248318_at |
AT5G52690
|
[Copper transport protein family] |
-0.717 |
-0.017 |
| 234 |
254481_at |
AT4G20480
|
[Putative endonuclease or glycosyl hydrolase] |
-0.708 |
0.008 |
| 235 |
259260_at |
AT3G11370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.707 |
-0.018 |
| 236 |
247645_at |
AT5G60530
|
[late embryogenesis abundant protein-related / LEA protein-related] |
-0.706 |
1.017 |
| 237 |
252511_at |
AT3G46280
|
[protein kinase-related] |
-0.693 |
1.005 |
| 238 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-0.687 |
0.073 |
| 239 |
267209_at |
AT2G30930
|
unknown |
-0.682 |
0.075 |
| 240 |
264640_at |
AT1G65680
|
ATEXPB2, expansin B2, ATHEXP BETA 1.4, EXPB2, expansin B2 |
-0.681 |
0.201 |
| 241 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
-0.680 |
0.055 |
| 242 |
266787_at |
AT2G28990
|
[Leucine-rich repeat protein kinase family protein] |
-0.677 |
-0.070 |
| 243 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.677 |
0.018 |
| 244 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.675 |
0.126 |
| 245 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.674 |
0.960 |
| 246 |
247933_at |
AT5G56980
|
unknown |
-0.671 |
0.128 |
| 247 |
252568_at |
AT3G45410
|
LecRK-I.3, L-type lectin receptor kinase I.3 |
-0.666 |
-0.061 |
| 248 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.664 |
0.037 |
| 249 |
266176_at |
AT2G02250
|
AtPP2-B2, phloem protein 2-B2, PP2-B2, phloem protein 2-B2 |
-0.661 |
-0.052 |
| 250 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
-0.660 |
0.462 |
| 251 |
251044_at |
AT5G02350
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.656 |
-0.099 |
| 252 |
249186_at |
AT5G43040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.650 |
0.370 |
| 253 |
260796_at |
AT1G78360
|
ATGSTU21, glutathione S-transferase TAU 21, GSTU21, glutathione S-transferase TAU 21 |
-0.649 |
-0.105 |
| 254 |
267240_at |
AT2G02680
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.646 |
0.356 |
| 255 |
251346_at |
AT3G60980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.644 |
0.145 |
| 256 |
266346_at |
AT2G01430
|
ATHB-17, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 17, ATHB17, homeobox-leucine zipper protein 17, HB17, homeobox-leucine zipper protein 17 |
-0.639 |
0.475 |
| 257 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
-0.638 |
0.212 |
| 258 |
261879_at |
AT1G50520
|
CYP705A27, cytochrome P450, family 705, subfamily A, polypeptide 27 |
-0.634 |
0.177 |
| 259 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.627 |
0.383 |
| 260 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
-0.626 |
0.176 |
| 261 |
256684_at |
AT3G32040
|
[Terpenoid synthases superfamily protein] |
-0.625 |
0.037 |
| 262 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.625 |
0.087 |
| 263 |
249557_at |
AT5G38280
|
PR5K, PR5-like receptor kinase |
-0.621 |
0.204 |
| 264 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.619 |
0.634 |
| 265 |
264101_at |
AT1G79000
|
ATHAC1, ARABIDOPSIS HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY 1, ATHPCAT2, ARABIDOPSIS THALIANA P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 2, HAC1, histone acetyltransferase of the CBP family 1, PCAT2, P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 2 |
-0.617 |
-0.034 |
| 266 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
-0.616 |
0.503 |
| 267 |
245106_at |
AT2G41650
|
unknown |
-0.610 |
0.171 |
| 268 |
258397_at |
AT3G15357
|
unknown |
-0.608 |
0.821 |
| 269 |
251107_at |
AT5G01610
|
unknown |
-0.608 |
0.262 |
| 270 |
267085_at |
AT2G32620
|
ATCSLB02, cellulose synthase-like B, ATCSLB2, CELLULOSE SYNTHASE LIKE B2, CSLB02, CELLULOSE SYNTHASE LIKE B2, CSLB02, cellulose synthase-like B |
-0.607 |
0.997 |
| 271 |
249185_at |
AT5G43030
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.606 |
0.459 |
| 272 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.606 |
0.026 |
| 273 |
263465_at |
AT2G31940
|
unknown |
-0.604 |
-0.112 |
| 274 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.602 |
0.339 |
| 275 |
255022_at |
AT4G10350
|
ANAC070, NAC domain containing protein 70, BRN2, BEARSKIN 2, NAC070, NAC domain containing protein 70 |
-0.602 |
-0.019 |
| 276 |
248079_at |
AT5G55790
|
unknown |
-0.601 |
0.019 |
| 277 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
-0.600 |
-0.035 |
| 278 |
250747_at |
AT5G05900
|
[UDP-Glycosyltransferase superfamily protein] |
-0.599 |
-0.084 |
| 279 |
267119_at |
AT2G32610
|
ATCSLB01, cellulose synthase-like B1, ATCSLB1, CELLULOSE SYNTHASE LIKE B1, CSLB01, CELLULOSE SYNTHASE LIKE B1, CSLB01, cellulose synthase-like B1 |
-0.597 |
1.255 |
| 280 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.597 |
-0.187 |
| 281 |
253132_at |
AT4G35580
|
CBNAC, calmodulin-binding NAC protein, NTL9, NAC transcription factor-like 9 |
-0.597 |
0.125 |
| 282 |
258005_at |
AT3G19390
|
[Granulin repeat cysteine protease family protein] |
-0.596 |
0.665 |
| 283 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
-0.594 |
0.262 |
| 284 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.591 |
0.402 |
| 285 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.590 |
0.828 |
| 286 |
256137_at |
AT1G48690
|
[Auxin-responsive GH3 family protein] |
-0.587 |
0.067 |
| 287 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
-0.587 |
0.311 |
| 288 |
258027_at |
AT3G19515
|
unknown |
-0.586 |
0.005 |
| 289 |
260611_at |
AT2G43670
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.585 |
0.122 |
| 290 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.584 |
0.097 |
| 291 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.583 |
-0.100 |
| 292 |
249562_at |
AT5G38350
|
[Disease resistance protein (NBS-LRR class) family] |
-0.581 |
-0.038 |
| 293 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.578 |
-0.628 |
| 294 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
-0.576 |
0.566 |
| 295 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
-0.575 |
0.309 |
| 296 |
256847_at |
AT3G27950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.574 |
0.068 |
| 297 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.572 |
0.432 |
| 298 |
257432_at |
AT2G21850
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.571 |
0.218 |
| 299 |
255730_at |
AT1G25460
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.571 |
-0.004 |
| 300 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
-0.569 |
0.169 |
| 301 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
-0.568 |
1.222 |
| 302 |
260752_at |
AT1G49030
|
[PLAC8 family protein] |
-0.565 |
-0.113 |
| 303 |
254912_at |
AT4G11230
|
[Riboflavin synthase-like superfamily protein] |
-0.559 |
-0.006 |
| 304 |
246807_at |
AT5G27100
|
ATGLR2.1, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 2.1, GLR2.1, glutamate receptor 2.1 |
-0.559 |
-0.099 |
| 305 |
262314_at |
AT1G70810
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.556 |
0.362 |
| 306 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
-0.556 |
0.479 |
| 307 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
-0.554 |
0.048 |
| 308 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
-0.553 |
0.387 |
| 309 |
250931_at |
AT5G03200
|
LUL1, LOG2-LIKE UBIQUITIN LIGASE1 |
-0.553 |
0.085 |
| 310 |
245547_at |
AT4G15300
|
CYP702A2, cytochrome P450, family 702, subfamily A, polypeptide 2 |
-0.552 |
-0.053 |
| 311 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.548 |
0.425 |
| 312 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.543 |
-0.296 |
| 313 |
254847_at |
AT4G11850
|
MEE54, maternal effect embryo arrest 54, PLDGAMMA1, phospholipase D gamma 1 |
-0.542 |
0.128 |
| 314 |
264835_at |
AT1G03550
|
AtSCAMP4, SCAMP4, Secretory carrier membrane protein 4 |
-0.541 |
0.011 |
| 315 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.540 |
0.572 |
| 316 |
259964_at |
AT1G53680
|
ATGSTU28, glutathione S-transferase TAU 28, GSTU28, glutathione S-transferase TAU 28 |
-0.537 |
-0.029 |
| 317 |
249766_at |
AT5G24070
|
[Peroxidase superfamily protein] |
-0.532 |
0.076 |
| 318 |
247591_at |
AT5G60770
|
ATNRT2.4, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.4, NRT2.4, nitrate transporter 2.4 |
-0.531 |
-0.010 |
| 319 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.530 |
0.572 |
| 320 |
266235_at |
AT2G02360
|
AtPP2-B10, phloem protein 2-B10, PP2-B10, phloem protein 2-B10 |
-0.526 |
0.037 |
| 321 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
-0.525 |
-0.120 |
| 322 |
252205_at |
AT3G50350
|
unknown |
-0.523 |
0.024 |
| 323 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.520 |
0.643 |
| 324 |
257154_at |
AT3G27210
|
unknown |
-0.515 |
0.386 |
| 325 |
255059_at |
AT4G09420
|
[Disease resistance protein (TIR-NBS class)] |
-0.515 |
0.246 |
| 326 |
252651_at |
AT3G44700
|
unknown |
-0.514 |
0.082 |
| 327 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.513 |
0.822 |
| 328 |
262206_at |
AT2G01090
|
[Ubiquinol-cytochrome C reductase hinge protein] |
-0.513 |
-0.062 |
| 329 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.512 |
-0.080 |
| 330 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.511 |
0.762 |
| 331 |
255924_at |
AT1G22170
|
[Phosphoglycerate mutase family protein] |
-0.509 |
0.176 |
| 332 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.508 |
0.153 |
| 333 |
249245_at |
AT5G42280
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.508 |
0.089 |
| 334 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
-0.506 |
0.091 |
| 335 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
-0.498 |
-0.228 |