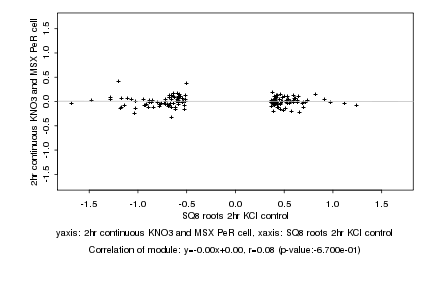
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
SQ8 roots 2hr KCl control |
Log2 signal ratio
2hr continuous KNO3 and MSX PeR cell |
| 1 |
248122_at |
AT5G54700
|
[Ankyrin repeat family protein] |
1.239 |
-0.065 |
| 2 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
1.117 |
-0.033 |
| 3 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.968 |
-0.006 |
| 4 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.906 |
0.052 |
| 5 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.811 |
0.159 |
| 6 |
251334_at |
AT3G61390
|
[RING/U-box superfamily protein] |
0.738 |
0.035 |
| 7 |
248052_at |
AT5G55800
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.717 |
-0.000 |
| 8 |
250135_at |
AT5G15360
|
unknown |
0.697 |
-0.000 |
| 9 |
254566_at |
AT4G19240
|
unknown |
0.693 |
-0.108 |
| 10 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.684 |
-0.039 |
| 11 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.654 |
-0.209 |
| 12 |
253796_at |
AT4G28460
|
unknown |
0.646 |
0.112 |
| 13 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.634 |
0.033 |
| 14 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.629 |
-0.015 |
| 15 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.612 |
0.074 |
| 16 |
255471_at |
AT4G03050
|
AOP3 |
0.603 |
0.069 |
| 17 |
252663_at |
AT3G44070
|
[Glycosyl hydrolase family 35 protein] |
0.595 |
0.031 |
| 18 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.592 |
0.134 |
| 19 |
245677_at |
AT1G56660
|
unknown |
0.587 |
-0.003 |
| 20 |
266282_at |
AT2G29220
|
LecRK-III.1, L-type lectin receptor kinase III.1 |
0.586 |
0.000 |
| 21 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
0.566 |
-0.204 |
| 22 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.560 |
0.011 |
| 23 |
254582_at |
AT4G19470
|
[Leucine-rich repeat (LRR) family protein] |
0.549 |
-0.033 |
| 24 |
265732_at |
AT2G01300
|
unknown |
0.539 |
0.053 |
| 25 |
260121_at |
AT1G33910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.532 |
0.032 |
| 26 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.526 |
0.026 |
| 27 |
260100_at |
AT1G73177
|
APC13, anaphase-promoting complex 13, BNS, BONSAI |
0.526 |
0.117 |
| 28 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
0.526 |
-0.023 |
| 29 |
251755_at |
AT3G55790
|
unknown |
0.522 |
0.037 |
| 30 |
245307_at |
AT4G16770
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.516 |
-0.020 |
| 31 |
250576_at |
AT5G08250
|
[Cytochrome P450 superfamily protein] |
0.511 |
-0.143 |
| 32 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.498 |
0.104 |
| 33 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.487 |
-0.171 |
| 34 |
253532_at |
AT4G31570
|
unknown |
0.479 |
0.019 |
| 35 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.474 |
0.083 |
| 36 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.468 |
0.010 |
| 37 |
255274_at |
AT4G05300
|
unknown |
0.464 |
0.017 |
| 38 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.462 |
-0.060 |
| 39 |
267135_at |
AT2G23430
|
ICK1, KRP1, KIP-RELATED PROTEIN 1 |
0.462 |
-0.156 |
| 40 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.460 |
0.150 |
| 41 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.459 |
-0.033 |
| 42 |
263617_at |
AT2G04670
|
[gypsy-like retrotransposon family, has a 1.2e-313 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
0.451 |
0.046 |
| 43 |
249169_at |
AT5G42880
|
unknown |
0.432 |
-0.122 |
| 44 |
253352_at |
AT4G33710
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.430 |
-0.122 |
| 45 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.427 |
0.141 |
| 46 |
265689_at |
AT2G24310
|
unknown |
0.425 |
-0.054 |
| 47 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.421 |
0.005 |
| 48 |
256859_at |
AT3G22940
|
[F-box associated ubiquitination effector family protein] |
0.421 |
-0.027 |
| 49 |
249634_at |
AT5G36840
|
unknown |
0.419 |
0.000 |
| 50 |
261967_at |
AT1G36440
|
unknown |
0.418 |
-0.014 |
| 51 |
259809_at |
AT1G49800
|
unknown |
0.418 |
-0.016 |
| 52 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.417 |
0.128 |
| 53 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.417 |
0.086 |
| 54 |
265186_at |
AT1G23560
|
unknown |
0.408 |
0.038 |
| 55 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
0.405 |
0.071 |
| 56 |
247047_at |
AT5G66650
|
unknown |
0.400 |
-0.017 |
| 57 |
264549_at |
AT1G09440
|
[Protein kinase superfamily protein] |
0.399 |
0.007 |
| 58 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
0.398 |
0.106 |
| 59 |
260845_at |
AT1G17310
|
[MADS-box transcription factor family protein] |
0.397 |
-0.057 |
| 60 |
258796_at |
AT3G04630
|
WDL1, WVD2-like 1 |
0.395 |
0.057 |
| 61 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.395 |
-0.017 |
| 62 |
259748_at |
AT1G71180
|
[6-phosphogluconate dehydrogenase family protein] |
0.394 |
0.018 |
| 63 |
266355_at |
AT2G01400
|
unknown |
0.394 |
0.061 |
| 64 |
248384_at |
AT5G51930
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.393 |
-0.068 |
| 65 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.384 |
-0.048 |
| 66 |
257321_at |
ATMG01130
|
ORF106F |
0.382 |
-0.190 |
| 67 |
256287_at |
AT3G12190
|
unknown |
0.381 |
-0.007 |
| 68 |
259979_at |
AT1G76600
|
unknown |
0.381 |
0.024 |
| 69 |
255881_at |
AT1G67070
|
DIN9, DARK INDUCIBLE 9, PMI2, PHOSPHOMANNOSE ISOMERASE 2 |
0.378 |
0.020 |
| 70 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.377 |
-0.040 |
| 71 |
245545_at |
AT4G15280
|
UGT71B5, UDP-glucosyl transferase 71B5 |
0.370 |
0.190 |
| 72 |
250162_at |
AT5G15250
|
ATFTSH6, FTSH6, FTSH protease 6 |
0.366 |
-0.095 |
| 73 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
0.365 |
-0.003 |
| 74 |
262821_at |
AT1G11800
|
[endonuclease/exonuclease/phosphatase family protein] |
0.365 |
0.025 |
| 75 |
254727_at |
AT4G13670
|
PTAC5, plastid transcriptionally active 5 |
0.361 |
-0.019 |
| 76 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
-1.689 |
-0.037 |
| 77 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
-1.486 |
0.038 |
| 78 |
264282_at |
AT1G61840
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.288 |
0.097 |
| 79 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
-1.282 |
0.056 |
| 80 |
266851_at |
AT2G26820
|
ATPP2-A3, phloem protein 2-A3, PP2-A3, phloem protein 2-A3 |
-1.207 |
0.423 |
| 81 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-1.188 |
-0.132 |
| 82 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
-1.176 |
-0.120 |
| 83 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
-1.174 |
0.073 |
| 84 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-1.142 |
-0.080 |
| 85 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
-1.109 |
0.066 |
| 86 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-1.074 |
0.053 |
| 87 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-1.036 |
-0.234 |
| 88 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
-1.033 |
-0.143 |
| 89 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
-1.030 |
0.017 |
| 90 |
246393_at |
AT1G58150
|
unknown |
-0.946 |
0.049 |
| 91 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.940 |
-0.081 |
| 92 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.932 |
-0.062 |
| 93 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.916 |
-0.059 |
| 94 |
265124_at |
AT1G55430
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.900 |
-0.118 |
| 95 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
-0.887 |
0.022 |
| 96 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.884 |
-0.015 |
| 97 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.867 |
-0.007 |
| 98 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.865 |
-0.000 |
| 99 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.859 |
0.027 |
| 100 |
254979_at |
AT4G10550
|
[Subtilase family protein] |
-0.850 |
-0.119 |
| 101 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
-0.806 |
-0.013 |
| 102 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
-0.787 |
-0.082 |
| 103 |
249617_at |
AT5G37450
|
[Leucine-rich repeat protein kinase family protein] |
-0.780 |
-0.043 |
| 104 |
248212_at |
AT5G54020
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.771 |
-0.046 |
| 105 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.767 |
-0.021 |
| 106 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.737 |
-0.038 |
| 107 |
252530_at |
AT3G46500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.726 |
0.006 |
| 108 |
255547_at |
AT4G01920
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.722 |
-0.046 |
| 109 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
-0.721 |
0.043 |
| 110 |
249185_at |
AT5G43030
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.707 |
-0.043 |
| 111 |
248415_at |
AT5G51620
|
[Uncharacterised protein family (UPF0172)] |
-0.694 |
-0.086 |
| 112 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.686 |
-0.079 |
| 113 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.686 |
0.098 |
| 114 |
264640_at |
AT1G65680
|
ATEXPB2, expansin B2, ATHEXP BETA 1.4, EXPB2, expansin B2 |
-0.683 |
0.084 |
| 115 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
-0.677 |
0.125 |
| 116 |
245674_at |
AT1G56680
|
[Chitinase family protein] |
-0.676 |
-0.035 |
| 117 |
252490_at |
AT3G46720
|
[UDP-Glycosyltransferase superfamily protein] |
-0.666 |
-0.314 |
| 118 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.665 |
0.042 |
| 119 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
-0.662 |
-0.115 |
| 120 |
245611_at |
AT4G14390
|
[Ankyrin repeat family protein] |
-0.658 |
0.129 |
| 121 |
262894_at |
AT1G59780
|
[NB-ARC domain-containing disease resistance protein] |
-0.644 |
-0.023 |
| 122 |
248416_at |
AT5G51630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.644 |
0.104 |
| 123 |
266176_at |
AT2G02250
|
AtPP2-B2, phloem protein 2-B2, PP2-B2, phloem protein 2-B2 |
-0.639 |
0.179 |
| 124 |
257432_at |
AT2G21850
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.629 |
0.088 |
| 125 |
257112_at |
AT3G20120
|
CYP705A21, cytochrome P450, family 705, subfamily A, polypeptide 21 |
-0.615 |
-0.123 |
| 126 |
252202_at |
AT3G50300
|
[HXXXD-type acyl-transferase family protein] |
-0.615 |
-0.157 |
| 127 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
-0.612 |
0.016 |
| 128 |
266838_at |
AT2G25980
|
[Mannose-binding lectin superfamily protein] |
-0.601 |
0.172 |
| 129 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
-0.601 |
0.044 |
| 130 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.601 |
0.020 |
| 131 |
249562_at |
AT5G38350
|
[Disease resistance protein (NBS-LRR class) family] |
-0.595 |
0.014 |
| 132 |
249897_at |
AT5G22550
|
unknown |
-0.594 |
-0.013 |
| 133 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
-0.594 |
0.107 |
| 134 |
248318_at |
AT5G52690
|
[Copper transport protein family] |
-0.594 |
0.038 |
| 135 |
248077_at |
AT5G55770
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.587 |
-0.054 |
| 136 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.587 |
0.072 |
| 137 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
-0.585 |
0.101 |
| 138 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
-0.581 |
0.162 |
| 139 |
245838_at |
AT1G58410
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.573 |
0.109 |
| 140 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.549 |
-0.017 |
| 141 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.547 |
-0.016 |
| 142 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
-0.545 |
0.073 |
| 143 |
254264_at |
AT4G23510
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.536 |
0.009 |
| 144 |
256684_at |
AT3G32040
|
[Terpenoid synthases superfamily protein] |
-0.532 |
-0.154 |
| 145 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.529 |
-0.076 |
| 146 |
247988_at |
AT5G56910
|
[Proteinase inhibitor I25, cystatin, conserved region] |
-0.521 |
0.013 |
| 147 |
258769_at |
AT3G10870
|
ATMES17, ARABIDOPSIS THALIANA METHYL ESTERASE 17, MES17, methyl esterase 17 |
-0.519 |
0.142 |
| 148 |
253132_at |
AT4G35580
|
CBNAC, calmodulin-binding NAC protein, NTL9, NAC transcription factor-like 9 |
-0.514 |
0.047 |
| 149 |
251934_at |
AT3G54070
|
[Ankyrin repeat family protein] |
-0.512 |
0.385 |