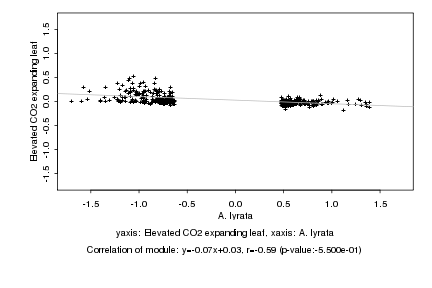
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
A. lyrata |
Log2 signal ratio
Elevated CO2 expanding leaf |
| 1 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
1.389 |
-0.005 |
| 2 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
1.387 |
-0.104 |
| 3 |
256460_at |
AT1G36240
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
1.356 |
-0.043 |
| 4 |
259054_at |
AT3G03480
|
CHAT, acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase |
1.350 |
-0.090 |
| 5 |
249598_at |
AT5G37970
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
1.349 |
-0.004 |
| 6 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
1.305 |
-0.068 |
| 7 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
1.290 |
0.026 |
| 8 |
250773_at |
AT5G05430
|
[RNA-binding protein] |
1.272 |
0.045 |
| 9 |
252639_at |
AT3G44550
|
FAR5, fatty acid reductase 5 |
1.245 |
-0.047 |
| 10 |
254699_at |
AT4G17990
|
unknown |
1.164 |
-0.054 |
| 11 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.152 |
0.025 |
| 12 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
1.112 |
-0.184 |
| 13 |
260930_at |
AT1G02620
|
[Ras-related small GTP-binding family protein] |
1.058 |
0.011 |
| 14 |
256338_at |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
1.007 |
0.046 |
| 15 |
260934_at |
AT1G45100
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
1.001 |
-0.008 |
| 16 |
265937_at |
AT2G19610
|
[RING/U-box superfamily protein] |
0.992 |
-0.034 |
| 17 |
255576_at |
AT4G01440
|
UMAMIT31, Usually multiple acids move in and out Transporters 31 |
0.956 |
-0.039 |
| 18 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.955 |
0.008 |
| 19 |
245814_at |
AT1G49910
|
BUB3.2, BUB (BUDDING UNINHIBITED BY BENZYMIDAZOL) 3.2 |
0.952 |
-0.038 |
| 20 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
0.946 |
-0.018 |
| 21 |
247932_at |
AT5G56920
|
[Cystatin/monellin superfamily protein] |
0.926 |
-0.007 |
| 22 |
257835_at |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
0.903 |
-0.048 |
| 23 |
264341_at |
AT1G70270
|
unknown |
0.891 |
-0.054 |
| 24 |
267569_at |
AT2G30790
|
PSBP-2, photosystem II subunit P-2 |
0.886 |
0.045 |
| 25 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.879 |
0.142 |
| 26 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.855 |
0.034 |
| 27 |
252677_at |
AT3G44320
|
AtNIT3, NITRILASE 3, NIT3, nitrilase 3 |
0.854 |
0.007 |
| 28 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
0.851 |
-0.037 |
| 29 |
258147_at |
AT3G18070
|
BGLU43, beta glucosidase 43 |
0.846 |
-0.021 |
| 30 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.840 |
0.025 |
| 31 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
0.834 |
-0.047 |
| 32 |
262464_at |
AT1G50280
|
[Phototropic-responsive NPH3 family protein] |
0.834 |
-0.079 |
| 33 |
261923_at |
AT1G22380
|
AtUGT85A3, UDP-glucosyl transferase 85A3, UGT85A3, UDP-glucosyl transferase 85A3 |
0.811 |
-0.071 |
| 34 |
257509_at |
AT1G63190
|
[Cystatin/monellin superfamily protein] |
0.810 |
-0.009 |
| 35 |
248691_at |
AT5G48310
|
unknown |
0.809 |
-0.100 |
| 36 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.804 |
0.021 |
| 37 |
246467_at |
AT5G17040
|
[UDP-Glycosyltransferase superfamily protein] |
0.803 |
-0.054 |
| 38 |
264181_at |
AT1G65350
|
UBQ13, ubiquitin 13 |
0.802 |
0.014 |
| 39 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.798 |
0.010 |
| 40 |
261025_at |
AT1G01225
|
[NC domain-containing protein-related] |
0.796 |
-0.068 |
| 41 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.793 |
-0.071 |
| 42 |
255756_at |
AT1G19940
|
AtGH9B5, glycosyl hydrolase 9B5, GH9B5, glycosyl hydrolase 9B5 |
0.788 |
0.018 |
| 43 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.784 |
-0.005 |
| 44 |
264987_at |
AT1G27030
|
unknown |
0.766 |
-0.064 |
| 45 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
0.763 |
-0.114 |
| 46 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.755 |
0.001 |
| 47 |
259353_at |
AT3G05190
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.754 |
-0.021 |
| 48 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.746 |
-0.042 |
| 49 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.728 |
-0.026 |
| 50 |
264853_at |
AT2G17260
|
ATGLR2, ATGLR3.1, GLR2, glutamate receptor 2, GLR3.1 |
0.722 |
-0.043 |
| 51 |
251512_at |
AT3G59190
|
[F-box/RNI-like superfamily protein] |
0.719 |
-0.041 |
| 52 |
245929_at |
AT5G24760
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.719 |
-0.019 |
| 53 |
265502_at |
AT2G15500
|
[RNA-binding protein] |
0.714 |
-0.037 |
| 54 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
0.713 |
0.022 |
| 55 |
246574_at |
AT1G31670
|
[Copper amine oxidase family protein] |
0.709 |
-0.043 |
| 56 |
253511_at |
AT4G31740
|
[Sec1/munc18-like (SM) proteins superfamily] |
0.702 |
-0.031 |
| 57 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.700 |
-0.039 |
| 58 |
265057_at |
AT1G52140
|
unknown |
0.697 |
0.035 |
| 59 |
266756_at |
AT2G46950
|
CYP709B2, cytochrome P450, family 709, subfamily B, polypeptide 2 |
0.689 |
-0.015 |
| 60 |
266305_at |
AT2G27120
|
POL2B, TIL2, TILTED 2 |
0.686 |
-0.012 |
| 61 |
258663_at |
AT3G08670
|
unknown |
0.674 |
0.020 |
| 62 |
250143_at |
AT5G14670
|
ARFA1B, ADP-ribosylation factor A1B, ATARFA1B, ADP-ribosylation factor A1B |
0.674 |
0.054 |
| 63 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
0.669 |
-0.047 |
| 64 |
248658_at |
AT5G48600
|
ATCAP-C, ARABIDOPSIS THALIANA CHROMOSOME ASSOCIATED PROTEIN-C, ATSMC3, structural maintenance of chromosome 3, ATSMC4, ARABIDOPSIS THALIANA STRUCTURAL MAINTENANCE OF CHROMOSOME 4, SMC3, structural maintenance of chromosome 3 |
0.669 |
-0.067 |
| 65 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
0.669 |
-0.008 |
| 66 |
255920_at |
AT5G28590
|
[DNA-binding family protein] |
0.661 |
-0.000 |
| 67 |
249995_at |
AT5G18610
|
[Protein kinase superfamily protein] |
0.657 |
0.046 |
| 68 |
257275_at |
AT3G14450
|
CID9, CTC-interacting domain 9 |
0.655 |
0.084 |
| 69 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.655 |
0.072 |
| 70 |
252769_at |
AT3G42850
|
[Mevalonate/galactokinase family protein] |
0.648 |
0.019 |
| 71 |
246030_at |
AT5G21105
|
[Plant L-ascorbate oxidase] |
0.643 |
0.014 |
| 72 |
248509_at |
AT5G50335
|
unknown |
0.637 |
0.098 |
| 73 |
260228_at |
AT1G74540
|
CYP98A8, cytochrome P450, family 98, subfamily A, polypeptide 8 |
0.630 |
-0.030 |
| 74 |
257825_at |
AT3G26700
|
[Protein kinase superfamily protein] |
0.628 |
-0.048 |
| 75 |
247682_at |
AT5G59650
|
[Leucine-rich repeat protein kinase family protein] |
0.627 |
0.051 |
| 76 |
251119_at |
AT3G63510
|
[FMN-linked oxidoreductases superfamily protein] |
0.616 |
-0.056 |
| 77 |
260109_at |
AT1G63260
|
TET10, tetraspanin10 |
0.613 |
0.001 |
| 78 |
261941_at |
AT1G22490
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.613 |
0.015 |
| 79 |
264066_at |
AT2G27880
|
AGO5, ARGONAUTE 5, AtAGO5 |
0.611 |
-0.037 |
| 80 |
263151_at |
AT1G54120
|
unknown |
0.608 |
-0.036 |
| 81 |
251473_at |
AT3G59610
|
[F-box family protein / jacalin lectin family protein] |
0.607 |
-0.038 |
| 82 |
266891_at |
AT2G44690
|
ARAC9, Arabidopsis RAC-like 9, ATROP8, RHO-RELATED PROTEIN FROM PLANTS 8, ROP8, RHO-RELATED PROTEIN FROM PLANTS 8 |
0.604 |
-0.045 |
| 83 |
265448_at |
AT2G46590
|
Dof-type zinc finger DNA-binding family protein |
0.603 |
0.025 |
| 84 |
263409_at |
AT2G04063
|
[glycine-rich protein] |
0.597 |
-0.011 |
| 85 |
257847_at |
AT3G13020
|
[hAT transposon superfamily protein] |
0.595 |
-0.079 |
| 86 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.594 |
-0.053 |
| 87 |
264861_at |
AT1G24320
|
[Six-hairpin glycosidases superfamily protein] |
0.592 |
-0.056 |
| 88 |
245141_at |
AT2G45400
|
BEN1, BRI1-5 ENHANCED 1 |
0.591 |
-0.061 |
| 89 |
260461_at |
AT1G10980
|
[Lung seven transmembrane receptor family protein] |
0.588 |
-0.023 |
| 90 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
0.582 |
-0.064 |
| 91 |
267465_at |
AT2G19170
|
SLP3, subtilisin-like serine protease 3 |
0.582 |
-0.046 |
| 92 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
0.579 |
-0.035 |
| 93 |
255417_at |
AT4G03190
|
AFB1, AUXIN SIGNALING F BOX PROTEIN 1, ATGRH1, GRH1, GRR1-like protein 1 |
0.577 |
0.005 |
| 94 |
250431_at |
AT5G10440
|
CYCD4;2, cyclin d4;2 |
0.576 |
0.023 |
| 95 |
246778_at |
AT5G27450
|
MK, mevalonate kinase, MVK, MEVALONATE KINASE |
0.573 |
-0.070 |
| 96 |
253010_at |
AT4G37750
|
ANT, AINTEGUMENTA, CKC, CKC1, COMPLEMENTING A PROTEIN KINASE C MUTANT 1, DRG, DRAGON |
0.572 |
-0.079 |
| 97 |
256355_at |
AT1G55040
|
[zinc finger (Ran-binding) family protein] |
0.567 |
-0.040 |
| 98 |
259637_at |
AT1G52260
|
ATPDI3, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 3, ATPDIL1-5, PDI-like 1-5, PDI3, PROTEIN DISULFIDE ISOMERASE 3, PDIL1-5, PDI-like 1-5 |
0.567 |
0.065 |
| 99 |
262106_at |
AT1G02970
|
ATWEE1, WEE1, WEE1 kinase homolog |
0.565 |
-0.049 |
| 100 |
261454_at |
AT1G21090
|
[Cupredoxin superfamily protein] |
0.563 |
-0.084 |
| 101 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.563 |
-0.022 |
| 102 |
246231_at |
AT4G37080
|
unknown |
0.561 |
-0.045 |
| 103 |
263430_at |
AT2G22270
|
unknown |
0.560 |
-0.023 |
| 104 |
256132_at |
AT1G13610
|
[alpha/beta-Hydrolases superfamily protein] |
0.559 |
0.019 |
| 105 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
0.559 |
0.010 |
| 106 |
256926_at |
AT3G22540
|
unknown |
0.557 |
-0.039 |
| 107 |
256626_at |
AT3G20015
|
[Eukaryotic aspartyl protease family protein] |
0.557 |
-0.025 |
| 108 |
257480_at |
AT1G15640
|
unknown |
0.556 |
-0.026 |
| 109 |
253721_at |
AT4G29250
|
[HXXXD-type acyl-transferase family protein] |
0.555 |
0.009 |
| 110 |
246906_at |
AT5G25475
|
[AP2/B3-like transcriptional factor family protein] |
0.554 |
-0.038 |
| 111 |
258630_at |
AT3G02820
|
[zinc knuckle (CCHC-type) family protein] |
0.553 |
-0.042 |
| 112 |
261129_at |
AT1G04820
|
TOR2, TORTIFOLIA 2, TUA4, tubulin alpha-4 chain |
0.550 |
-0.020 |
| 113 |
251229_at |
AT3G62740
|
BGLU7, beta glucosidase 7 |
0.549 |
-0.001 |
| 114 |
265005_at |
AT1G61667
|
unknown |
0.548 |
0.107 |
| 115 |
254140_at |
AT4G24610
|
unknown |
0.545 |
-0.103 |
| 116 |
250653_at |
AT5G06930
|
[LOCATED IN: chloroplast] |
0.544 |
0.026 |
| 117 |
259608_at |
AT1G27960
|
ECT9, evolutionarily conserved C-terminal region 9 |
0.544 |
-0.066 |
| 118 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
0.538 |
0.020 |
| 119 |
249718_at |
AT5G35740
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.537 |
-0.035 |
| 120 |
246155_at |
AT5G20030
|
[Plant Tudor-like RNA-binding protein] |
0.536 |
-0.030 |
| 121 |
257065_at |
AT3G18220
|
LPP4, lipid phosphate phosphatase 4 |
0.535 |
-0.048 |
| 122 |
245903_at |
AT5G11100
|
ATSYTD, NTMC2T2.2, NTMC2TYPE2.2, SYT4, synaptotagmin 4, SYTD |
0.534 |
0.011 |
| 123 |
249261_at |
AT5G41710
|
[Mutator-like transposase family, has a 6.2e-18 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.534 |
-0.047 |
| 124 |
256704_at |
AT3G30280
|
[HXXXD-type acyl-transferase family protein] |
0.534 |
0.016 |
| 125 |
252439_at |
AT3G47400
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.534 |
-0.022 |
| 126 |
253354_at |
AT4G33370
|
[DEA(D/H)-box RNA helicase family protein] |
0.532 |
-0.048 |
| 127 |
250469_at |
AT5G10130
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.530 |
-0.058 |
| 128 |
267547_at |
AT2G32670
|
ATVAMP725, vesicle-associated membrane protein 725, VAMP725, vesicle-associated membrane protein 725 |
0.529 |
-0.030 |
| 129 |
251881_at |
AT3G54250
|
[GHMP kinase family protein] |
0.528 |
-0.064 |
| 130 |
257417_at |
AT1G10110
|
[F-box family protein] |
0.523 |
-0.020 |
| 131 |
258241_at |
AT3G27650
|
LBD25, LOB domain-containing protein 25 |
0.522 |
0.004 |
| 132 |
252279_at |
AT3G49700
|
ACS9, 1-aminocyclopropane-1-carboxylate synthase 9, AtACS9, ETO3, ETHYLENE OVERPRODUCING 3 |
0.522 |
0.051 |
| 133 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.519 |
-0.146 |
| 134 |
250448_at |
AT5G10820
|
[Major facilitator superfamily protein] |
0.518 |
-0.009 |
| 135 |
264473_at |
AT1G67180
|
[zinc finger (C3HC4-type RING finger) family protein / BRCT domain-containing protein] |
0.517 |
-0.029 |
| 136 |
265382_at |
AT2G16790
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.516 |
0.022 |
| 137 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.514 |
0.011 |
| 138 |
260242_at |
AT1G63650
|
AtEGL3, ATMYC-2, EGL1, EGL3, ENHANCER OF GLABRA 3 |
0.514 |
-0.072 |
| 139 |
265432_at |
AT2G20650
|
FLY2, FLYING SAUCER 2 |
0.513 |
-0.035 |
| 140 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
0.512 |
-0.025 |
| 141 |
253891_at |
AT4G27720
|
[Major facilitator superfamily protein] |
0.511 |
-0.026 |
| 142 |
257493_at |
AT1G48180
|
unknown |
0.510 |
-0.039 |
| 143 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.510 |
-0.122 |
| 144 |
251236_at |
AT3G62380
|
unknown |
0.509 |
-0.004 |
| 145 |
262920_at |
AT1G79500
|
AtkdsA1 |
0.508 |
-0.052 |
| 146 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.508 |
-0.004 |
| 147 |
255536_at |
AT4G01575
|
[serine protease inhibitor, Kazal-type family protein] |
0.508 |
-0.041 |
| 148 |
258951_at |
AT3G01380
|
[transferases] |
0.508 |
-0.012 |
| 149 |
253195_at |
AT4G35420
|
DRL1, dihydroflavonol 4-reductase-like1, TKPR1, tetraketide alpha-pyrone reductase 1 |
0.507 |
-0.058 |
| 150 |
259376_at |
AT3G16320
|
Tetratricopeptide repeat (TPR)-like superfamily protein |
0.507 |
-0.026 |
| 151 |
250637_at |
AT5G07530
|
ATGRP-7, ARABIDOPSIS THALIANA GLYCINE RICH PROTEIN 7, ATGRP17, ARABIDOPSIS THALIANA GLYCINE RICH PROTEIN 17, GRP17, glycine rich protein 17 |
0.507 |
0.003 |
| 152 |
264939_at |
AT1G60630
|
[Leucine-rich repeat protein kinase family protein] |
0.505 |
-0.045 |
| 153 |
245113_at |
AT2G41660
|
MIZ1, mizu-kussei 1 |
0.505 |
0.002 |
| 154 |
253909_at |
AT4G27270
|
[Quinone reductase family protein] |
0.504 |
0.034 |
| 155 |
264296_at |
AT1G78720
|
[SecY protein transport family protein] |
0.504 |
-0.049 |
| 156 |
264990_at |
AT1G27210
|
[ARM repeat superfamily protein] |
0.503 |
-0.010 |
| 157 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
0.500 |
-0.027 |
| 158 |
254005_at |
AT4G26330
|
ATSBT3.18, UNE17, UNFERTILIZED EMBRYO SAC 17 |
0.499 |
-0.030 |
| 159 |
245902_at |
AT5G11080
|
[Ubiquitin-like superfamily protein] |
0.497 |
0.025 |
| 160 |
250717_at |
AT5G06200
|
CASP4, Casparian strip membrane domain protein 4 |
0.497 |
-0.020 |
| 161 |
263047_at |
AT2G17630
|
PSAT2, phosphoserine aminotransferase 2 |
0.496 |
-0.075 |
| 162 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.496 |
-0.091 |
| 163 |
249362_at |
AT5G40550
|
AtSGF29b, SGF29b, SaGa associated Factor 29 b |
0.496 |
-0.035 |
| 164 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
0.495 |
0.033 |
| 165 |
257342_at |
AT1G42365
|
[gypsy-like retrotransposon family (Athila), has a 9.0e-234 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.494 |
-0.019 |
| 166 |
261764_at |
AT1G15530
|
LecRK-S.1, L-type lectin receptor kinase S.1 |
0.493 |
0.002 |
| 167 |
251702_at |
AT3G56660
|
BZIP49, basic region/leucine zipper motif protein 49 |
0.492 |
0.010 |
| 168 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
0.490 |
-0.003 |
| 169 |
260389_at |
AT1G74055
|
unknown |
0.489 |
-0.051 |
| 170 |
255815_at |
AT1G19890
|
ATMGH3, MALE-GAMETE-SPECIFIC HISTONE H3, MGH3, male-gamete-specific histone H3 |
0.489 |
-0.007 |
| 171 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
0.488 |
-0.048 |
| 172 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
0.488 |
-0.040 |
| 173 |
265425_at |
AT2G20770
|
GCL2, GCR2-like 2 |
0.485 |
-0.020 |
| 174 |
261239_at |
AT1G32930
|
AtGALT31A, GALT31A, glycosyltransferase of CAZY family GT31 A |
0.484 |
-0.054 |
| 175 |
255531_at |
AT4G02160
|
unknown |
0.482 |
0.034 |
| 176 |
250630_at |
AT5G07400
|
[forkhead-associated domain-containing protein / FHA domain-containing protein] |
0.482 |
0.034 |
| 177 |
259263_at |
AT3G01010
|
[UDP-glucose/GDP-mannose dehydrogenase family protein] |
0.481 |
0.040 |
| 178 |
255258_at |
AT4G05060
|
[PapD-like superfamily protein] |
0.479 |
0.015 |
| 179 |
249954_at |
AT5G18920
|
[Cox19-like CHCH family protein] |
0.478 |
0.002 |
| 180 |
266913_at |
AT2G45890
|
ATROPGEF4, RHO GUANYL-NUCLEOTIDE EXCHANGE FACTOR 4, GEF4, guanine exchange factor 4, RHS11, ROOT HAIR SPECIFIC 11, ROPGEF4, ROP (rho of plants) guanine nucleotide exchange factor 4 |
0.478 |
-0.099 |
| 181 |
245743_at |
AT1G51080
|
unknown |
0.478 |
-0.080 |
| 182 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.476 |
-0.001 |
| 183 |
265179_at |
AT1G23650
|
unknown |
0.476 |
-0.067 |
| 184 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
0.476 |
0.019 |
| 185 |
257647_at |
AT3G25805
|
unknown |
0.475 |
-0.039 |
| 186 |
257473_at |
AT1G33840
|
unknown |
0.475 |
0.089 |
| 187 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
0.474 |
-0.040 |
| 188 |
263365_at |
AT2G20550
|
[HSP40/DnaJ peptide-binding protein] |
0.473 |
0.037 |
| 189 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
0.472 |
0.006 |
| 190 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-1.708 |
0.014 |
| 191 |
249482_at |
AT5G38980
|
unknown |
-1.603 |
0.010 |
| 192 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-1.578 |
0.306 |
| 193 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.535 |
0.059 |
| 194 |
256442_at |
AT3G10930
|
unknown |
-1.517 |
0.220 |
| 195 |
258114_at |
AT3G14660
|
CYP72A13, cytochrome P450, family 72, subfamily A, polypeptide 13 |
-1.410 |
0.004 |
| 196 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.404 |
0.022 |
| 197 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-1.361 |
0.104 |
| 198 |
263875_at |
AT2G21970
|
SEP2, stress enhanced protein 2 |
-1.357 |
0.018 |
| 199 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-1.354 |
0.301 |
| 200 |
252462_at |
AT3G47250
|
unknown |
-1.310 |
0.030 |
| 201 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
-1.256 |
0.084 |
| 202 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-1.228 |
0.057 |
| 203 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-1.225 |
0.058 |
| 204 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-1.224 |
0.379 |
| 205 |
265611_at |
AT2G25510
|
unknown |
-1.222 |
0.005 |
| 206 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-1.222 |
0.037 |
| 207 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-1.203 |
0.264 |
| 208 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-1.201 |
0.141 |
| 209 |
252561_at |
AT3G45980
|
H2B, HISTONE H2B, HTB9 |
-1.196 |
-0.010 |
| 210 |
248415_at |
AT5G51620
|
[Uncharacterised protein family (UPF0172)] |
-1.195 |
0.018 |
| 211 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-1.180 |
0.353 |
| 212 |
254040_at |
AT4G25900
|
[Galactose mutarotase-like superfamily protein] |
-1.179 |
0.087 |
| 213 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
-1.172 |
0.030 |
| 214 |
254248_at |
AT4G23270
|
CRK19, cysteine-rich RLK (RECEPTOR-like protein kinase) 19 |
-1.158 |
0.097 |
| 215 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-1.148 |
0.225 |
| 216 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
-1.143 |
0.015 |
| 217 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-1.143 |
0.099 |
| 218 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
-1.131 |
0.235 |
| 219 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
-1.119 |
0.444 |
| 220 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-1.113 |
0.181 |
| 221 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.103 |
0.489 |
| 222 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
-1.096 |
0.113 |
| 223 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
-1.095 |
0.198 |
| 224 |
262876_at |
AT1G64750
|
ATDSS1(I), deletion of SUV3 suppressor 1(I), DSS1(I), deletion of SUV3 suppressor 1(I) |
-1.094 |
-0.006 |
| 225 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-1.090 |
0.226 |
| 226 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-1.089 |
0.281 |
| 227 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-1.083 |
0.021 |
| 228 |
259385_at |
AT1G13470
|
unknown |
-1.075 |
0.388 |
| 229 |
257771_at |
AT3G23000
|
ATSR2, ATSRPK1, CIPK7, CBL-interacting protein kinase 7, PKS7, SnRK3.10, SNF1-RELATED PROTEIN KINASE 3.10 |
-1.074 |
0.051 |
| 230 |
260005_at |
AT1G67920
|
unknown |
-1.063 |
0.241 |
| 231 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
-1.062 |
0.529 |
| 232 |
258027_at |
AT3G19515
|
unknown |
-1.061 |
-0.014 |
| 233 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
-1.060 |
0.291 |
| 234 |
254955_at |
AT4G10920
|
KELP |
-1.046 |
0.021 |
| 235 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-1.044 |
0.167 |
| 236 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-1.042 |
0.037 |
| 237 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-1.035 |
0.192 |
| 238 |
245765_at |
AT1G33600
|
[Leucine-rich repeat (LRR) family protein] |
-1.032 |
0.009 |
| 239 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-1.032 |
0.143 |
| 240 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-1.031 |
0.224 |
| 241 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-1.022 |
0.174 |
| 242 |
263182_at |
AT1G05575
|
unknown |
-1.008 |
0.163 |
| 243 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-1.001 |
0.183 |
| 244 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.000 |
0.317 |
| 245 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.997 |
0.150 |
| 246 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.993 |
0.376 |
| 247 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.993 |
-0.006 |
| 248 |
259752_at |
AT1G71040
|
LPR2, Low Phosphate Root2 |
-0.982 |
0.072 |
| 249 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
-0.973 |
0.174 |
| 250 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.968 |
0.102 |
| 251 |
256754_at |
AT3G25690
|
AtCHUP1, Arabidopsis thaliana CHLOROPLAST UNUSUAL POSITIONING 1, CHUP1, CHLOROPLAST UNUSUAL POSITIONING 1 |
-0.967 |
-0.005 |
| 252 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-0.964 |
0.069 |
| 253 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.964 |
0.018 |
| 254 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.962 |
0.145 |
| 255 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
-0.961 |
0.223 |
| 256 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.961 |
0.092 |
| 257 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.957 |
0.039 |
| 258 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.956 |
0.021 |
| 259 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-0.954 |
0.401 |
| 260 |
260031_at |
AT1G68790
|
CRWN3, CROWDED NUCLEI 3, LINC3, LITTLE NUCLEI3 |
-0.953 |
-0.039 |
| 261 |
259479_at |
AT1G19020
|
unknown |
-0.946 |
0.215 |
| 262 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
-0.942 |
0.212 |
| 263 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
-0.942 |
0.332 |
| 264 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
-0.936 |
0.008 |
| 265 |
258000_at |
AT3G28940
|
[AIG2-like (avirulence induced gene) family protein] |
-0.931 |
0.051 |
| 266 |
256050_at |
AT1G07000
|
ATEXO70B2, exocyst subunit exo70 family protein B2, EXO70B2, exocyst subunit exo70 family protein B2 |
-0.929 |
0.144 |
| 267 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.928 |
0.006 |
| 268 |
253135_at |
AT4G35830
|
ACO1, aconitase 1 |
-0.928 |
0.002 |
| 269 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.912 |
-0.039 |
| 270 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
-0.902 |
0.248 |
| 271 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
-0.888 |
0.199 |
| 272 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
-0.883 |
-0.044 |
| 273 |
255504_at |
AT4G02200
|
[Drought-responsive family protein] |
-0.874 |
-0.006 |
| 274 |
266480_at |
AT2G31130
|
unknown |
-0.870 |
-0.003 |
| 275 |
263083_at |
AT2G27190
|
ATPAP1, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 1, ATPAP12, PURPLE ACID PHOSPHATASE 12, PAP1, PURPLE ACID PHOSPHATASE 1, PAP12, purple acid phosphatase 12 |
-0.865 |
0.055 |
| 276 |
266259_at |
AT2G27830
|
unknown |
-0.861 |
0.123 |
| 277 |
261317_at |
AT1G53030
|
[Cytochrome C oxidase copper chaperone (COX17)] |
-0.852 |
0.038 |
| 278 |
247800_at |
AT5G58570
|
unknown |
-0.852 |
0.173 |
| 279 |
258092_at |
AT3G14595
|
[Ribosomal protein L18ae family] |
-0.846 |
0.018 |
| 280 |
258203_at |
AT3G13950
|
unknown |
-0.845 |
0.256 |
| 281 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.843 |
0.394 |
| 282 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.838 |
0.499 |
| 283 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
-0.833 |
0.244 |
| 284 |
253593_at |
AT4G30760
|
[Putative endonuclease or glycosyl hydrolase] |
-0.830 |
-0.016 |
| 285 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
-0.828 |
0.181 |
| 286 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.827 |
0.231 |
| 287 |
258049_at |
AT3G16220
|
[Predicted eukaryotic LigT] |
-0.821 |
0.026 |
| 288 |
254894_at |
AT4G11840
|
PLDGAMMA3, phospholipase D gamma 3 |
-0.820 |
0.051 |
| 289 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-0.820 |
0.004 |
| 290 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
-0.819 |
0.212 |
| 291 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.818 |
0.094 |
| 292 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
-0.815 |
0.127 |
| 293 |
266658_at |
AT2G25735
|
unknown |
-0.810 |
0.072 |
| 294 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.810 |
0.059 |
| 295 |
248568_at |
AT5G49760
|
[Leucine-rich repeat protein kinase family protein] |
-0.809 |
0.064 |
| 296 |
252073_at |
AT3G51750
|
unknown |
-0.809 |
0.011 |
| 297 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.807 |
0.207 |
| 298 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.806 |
0.024 |
| 299 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.803 |
0.006 |
| 300 |
245384_at |
AT4G16790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.798 |
0.094 |
| 301 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.797 |
0.143 |
| 302 |
250395_at |
AT5G10950
|
[Tudor/PWWP/MBT superfamily protein] |
-0.792 |
-0.039 |
| 303 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.790 |
0.264 |
| 304 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.790 |
0.222 |
| 305 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
-0.785 |
-0.041 |
| 306 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.784 |
0.054 |
| 307 |
255037_at |
AT4G09460
|
AtMYB6, myb domain protein 6, MYB6, myb domain protein 6 |
-0.778 |
0.029 |
| 308 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.776 |
0.210 |
| 309 |
255154_at |
AT4G08220
|
[Mutator-like transposase family, has a 5.3e-67 P-value blast match to Q9SUF8 /145-308 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-0.776 |
-0.026 |
| 310 |
258999_at |
AT3G01850
|
[Aldolase-type TIM barrel family protein] |
-0.775 |
0.021 |
| 311 |
263759_at |
AT2G21290
|
unknown |
-0.772 |
-0.014 |
| 312 |
247426_at |
AT5G62570
|
CBP60a, calmodulin-binding protein 60a |
-0.772 |
0.060 |
| 313 |
264368_at |
AT1G03280
|
[Transcription factor TFIIE, alpha subunit] |
-0.765 |
0.009 |
| 314 |
265960_at |
AT2G37470
|
[Histone superfamily protein] |
-0.761 |
-0.031 |
| 315 |
261212_at |
AT1G12880
|
atnudt12, nudix hydrolase homolog 12, NUDT12, nudix hydrolase homolog 12 |
-0.761 |
0.068 |
| 316 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.760 |
0.039 |
| 317 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.758 |
0.048 |
| 318 |
261221_at |
AT1G19960
|
unknown |
-0.757 |
0.002 |
| 319 |
245729_at |
AT1G73490
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.753 |
-0.021 |
| 320 |
256603_at |
AT3G28270
|
unknown |
-0.748 |
-0.045 |
| 321 |
261193_at |
AT1G32920
|
unknown |
-0.744 |
0.027 |
| 322 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
-0.744 |
0.117 |
| 323 |
257591_at |
AT3G24900
|
AtRLP39, receptor like protein 39, RLP39, receptor like protein 39 |
-0.741 |
0.180 |
| 324 |
249622_at |
AT5G37550
|
unknown |
-0.741 |
-0.062 |
| 325 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-0.730 |
0.031 |
| 326 |
252165_at |
AT3G50550
|
unknown |
-0.730 |
0.053 |
| 327 |
256216_at |
AT1G56340
|
AtCRT1a, CRT1, calreticulin 1, CRT1a, calreticulin 1a |
-0.730 |
-0.001 |
| 328 |
251987_at |
AT3G53280
|
CYP71B5, cytochrome p450 71b5 |
-0.728 |
0.002 |
| 329 |
252093_at |
AT3G51500
|
unknown |
-0.727 |
0.039 |
| 330 |
248889_at |
AT5G46230
|
unknown |
-0.726 |
0.101 |
| 331 |
249839_at |
AT5G23405
|
[HMG-box (high mobility group) DNA-binding family protein] |
-0.725 |
-0.028 |
| 332 |
260155_at |
AT1G52870
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.722 |
0.011 |
| 333 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
-0.718 |
0.030 |
| 334 |
261609_at |
AT1G49740
|
[PLC-like phosphodiesterases superfamily protein] |
-0.712 |
-0.022 |
| 335 |
250689_at |
AT5G06610
|
unknown |
-0.709 |
0.072 |
| 336 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-0.708 |
0.025 |
| 337 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
-0.706 |
0.014 |
| 338 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
-0.706 |
0.092 |
| 339 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.705 |
0.069 |
| 340 |
253780_at |
AT4G28400
|
[Protein phosphatase 2C family protein] |
-0.704 |
0.092 |
| 341 |
254526_at |
AT4G19600
|
CYCT1;4 |
-0.703 |
0.030 |
| 342 |
260592_at |
AT1G55850
|
ATCSLE1, CSLE1, cellulose synthase like E1 |
-0.701 |
0.024 |
| 343 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
-0.700 |
0.036 |
| 344 |
261445_at |
AT1G28380
|
NSL1, necrotic spotted lesions 1 |
-0.699 |
0.011 |
| 345 |
256617_at |
AT3G22240
|
unknown |
-0.699 |
0.017 |
| 346 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
-0.698 |
0.017 |
| 347 |
249322_at |
AT5G40930
|
TOM20-4, translocase of outer membrane 20-4 |
-0.696 |
0.004 |
| 348 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-0.690 |
0.214 |
| 349 |
250728_at |
AT5G06440
|
unknown |
-0.689 |
0.030 |
| 350 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.689 |
-0.010 |
| 351 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
-0.686 |
0.131 |
| 352 |
264673_at |
AT1G09795
|
ATATP-PRT2, ATP phosphoribosyl transferase 2, ATP-PRT2, ATP phosphoribosyl transferase 2, HISN1B |
-0.685 |
-0.027 |
| 353 |
262057_at |
AT1G80040
|
unknown |
-0.684 |
-0.000 |
| 354 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.684 |
-0.012 |
| 355 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.682 |
0.168 |
| 356 |
263428_at |
AT2G22310
|
ATUBP4, ubiquitin-specific protease 4, UBP4, ubiquitin-specific protease 4 |
-0.681 |
0.014 |
| 357 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
-0.680 |
0.305 |
| 358 |
260012_at |
AT1G67865
|
unknown |
-0.675 |
0.068 |
| 359 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
-0.675 |
0.111 |
| 360 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.674 |
-0.074 |
| 361 |
258587_at |
AT3G04310
|
unknown |
-0.674 |
-0.021 |
| 362 |
262346_at |
AT1G63980
|
[D111/G-patch domain-containing protein] |
-0.673 |
-0.010 |
| 363 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.670 |
0.018 |
| 364 |
261294_at |
AT1G48430
|
[Dihydroxyacetone kinase] |
-0.665 |
0.067 |
| 365 |
247063_at |
AT5G66820
|
unknown |
-0.664 |
0.003 |
| 366 |
263654_at |
AT1G04300
|
[TRAF-like superfamily protein] |
-0.662 |
0.010 |
| 367 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.660 |
0.113 |
| 368 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.660 |
0.188 |
| 369 |
255283_at |
AT4G04620
|
ATG8B, autophagy 8b |
-0.660 |
0.031 |
| 370 |
263621_at |
AT2G04630
|
NRPB6B, NRPE6B |
-0.656 |
-0.010 |
| 371 |
247398_at |
AT5G62950
|
[RNA polymerase II, Rpb4, core protein] |
-0.652 |
0.023 |
| 372 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.651 |
-0.043 |
| 373 |
248923_at |
AT5G45940
|
AtNUDT11, Arabidopsis thaliana nudix hydrolase homolog 11, AtNUDX11, Arabidopsis thaliana nudix hydrolase homolog 11, NUDT11, nudix hydrolase homolog 11, NUDX11, nudix hydrolase homolog 11 |
-0.646 |
0.005 |
| 374 |
254481_at |
AT4G20480
|
[Putative endonuclease or glycosyl hydrolase] |
-0.643 |
-0.006 |
| 375 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
-0.642 |
-0.042 |
| 376 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
-0.642 |
0.005 |
| 377 |
262605_at |
AT1G15170
|
[MATE efflux family protein] |
-0.640 |
0.030 |