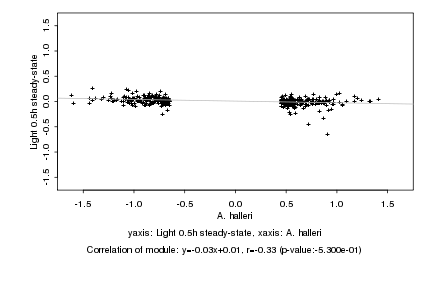
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
A. halleri |
Log2 signal ratio
Light 0.5h steady-state |
| 1 |
256460_at |
AT1G36240
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
1.407 |
0.058 |
| 2 |
253658_at |
AT4G30120
|
ATHMA3, A. THALIANA HEAVY METAL ATPASE 3, HMA3, heavy metal atpase 3 |
1.330 |
0.016 |
| 3 |
250773_at |
AT5G05430
|
[RNA-binding protein] |
1.320 |
0.008 |
| 4 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
1.240 |
0.030 |
| 5 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
1.200 |
0.068 |
| 6 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
1.170 |
0.105 |
| 7 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.167 |
0.017 |
| 8 |
264066_at |
AT2G27880
|
AGO5, ARGONAUTE 5, AtAGO5 |
1.086 |
0.019 |
| 9 |
263217_at |
AT1G30740
|
[FAD-binding Berberine family protein] |
1.049 |
-0.066 |
| 10 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
1.047 |
-0.056 |
| 11 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
1.020 |
0.000 |
| 12 |
264181_at |
AT1G65350
|
UBQ13, ubiquitin 13 |
1.016 |
0.168 |
| 13 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
0.995 |
0.144 |
| 14 |
251949_at |
AT3G53590
|
[Leucine-rich repeat protein kinase family protein] |
0.962 |
-0.042 |
| 15 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.961 |
-0.012 |
| 16 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
0.961 |
0.043 |
| 17 |
261217_at |
AT1G32850
|
UBP11, ubiquitin-specific protease 11 |
0.948 |
-0.054 |
| 18 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
0.944 |
-0.149 |
| 19 |
260930_at |
AT1G02620
|
[Ras-related small GTP-binding family protein] |
0.924 |
0.023 |
| 20 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
0.923 |
-0.017 |
| 21 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.921 |
0.028 |
| 22 |
262350_at |
AT2G48150
|
ATGPX4, glutathione peroxidase 4, GPX4, glutathione peroxidase 4 |
0.909 |
0.012 |
| 23 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.908 |
-0.162 |
| 24 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.901 |
-0.647 |
| 25 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
0.895 |
-0.077 |
| 26 |
254647_at |
AT4G18540
|
unknown |
0.888 |
0.025 |
| 27 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.884 |
0.013 |
| 28 |
260649_at |
AT1G08080
|
ACA7, alpha carbonic anhydrase 7, ATACA7, A. THALIANA ALPHA CARBONIC ANHYDRASE 7 |
0.879 |
0.089 |
| 29 |
251512_at |
AT3G59190
|
[F-box/RNI-like superfamily protein] |
0.868 |
-0.036 |
| 30 |
267569_at |
AT2G30790
|
PSBP-2, photosystem II subunit P-2 |
0.864 |
-0.039 |
| 31 |
262416_at |
AT1G49390
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.863 |
0.010 |
| 32 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.862 |
-0.320 |
| 33 |
252677_at |
AT3G44320
|
AtNIT3, NITRILASE 3, NIT3, nitrilase 3 |
0.858 |
0.004 |
| 34 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.844 |
-0.021 |
| 35 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.826 |
-0.190 |
| 36 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.825 |
0.025 |
| 37 |
265587_at |
AT2G19980
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.825 |
-0.035 |
| 38 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
0.821 |
0.088 |
| 39 |
253754_at |
AT4G29020
|
[glycine-rich protein] |
0.819 |
0.016 |
| 40 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
0.818 |
-0.017 |
| 41 |
257835_at |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
0.815 |
0.015 |
| 42 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
0.812 |
-0.030 |
| 43 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.799 |
-0.031 |
| 44 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.792 |
0.042 |
| 45 |
267622_at |
AT2G39690
|
unknown |
0.787 |
-0.048 |
| 46 |
250588_at |
AT5G07660
|
SMC6A, structural maintenance of chromosomes 6A |
0.786 |
-0.003 |
| 47 |
256450_at |
AT1G75290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.767 |
0.056 |
| 48 |
264235_at |
AT1G54560
|
ATXIE, MYOSIN XI E, XIE |
0.764 |
0.155 |
| 49 |
248658_at |
AT5G48600
|
ATCAP-C, ARABIDOPSIS THALIANA CHROMOSOME ASSOCIATED PROTEIN-C, ATSMC3, structural maintenance of chromosome 3, ATSMC4, ARABIDOPSIS THALIANA STRUCTURAL MAINTENANCE OF CHROMOSOME 4, SMC3, structural maintenance of chromosome 3 |
0.762 |
-0.006 |
| 50 |
249995_at |
AT5G18610
|
[Protein kinase superfamily protein] |
0.757 |
0.047 |
| 51 |
252530_at |
AT3G46500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.757 |
0.018 |
| 52 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.755 |
-0.046 |
| 53 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
0.754 |
-0.012 |
| 54 |
266756_at |
AT2G46950
|
CYP709B2, cytochrome P450, family 709, subfamily B, polypeptide 2 |
0.740 |
-0.015 |
| 55 |
257839_at |
AT3G25080
|
unknown |
0.734 |
-0.009 |
| 56 |
249954_at |
AT5G18920
|
[Cox19-like CHCH family protein] |
0.721 |
0.056 |
| 57 |
249461_at |
AT5G39640
|
[Putative endonuclease or glycosyl hydrolase] |
0.720 |
0.043 |
| 58 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.719 |
-0.443 |
| 59 |
265502_at |
AT2G15500
|
[RNA-binding protein] |
0.718 |
0.034 |
| 60 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
0.718 |
0.054 |
| 61 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.716 |
-0.070 |
| 62 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
0.708 |
0.046 |
| 63 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.703 |
0.026 |
| 64 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.697 |
0.037 |
| 65 |
259691_at |
AT1G63200
|
[Cystatin/monellin superfamily protein] |
0.693 |
-0.043 |
| 66 |
247170_at |
AT5G65530
|
[Protein kinase superfamily protein] |
0.684 |
0.002 |
| 67 |
257473_at |
AT1G33840
|
unknown |
0.682 |
-0.054 |
| 68 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
0.682 |
-0.087 |
| 69 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
0.681 |
0.101 |
| 70 |
250327_at |
AT5G12050
|
unknown |
0.670 |
-0.120 |
| 71 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.670 |
-0.001 |
| 72 |
252107_at |
AT3G51490
|
TMT3, tonoplast monosaccharide transporter3 |
0.656 |
0.006 |
| 73 |
263730_at |
AT1G60090
|
BGLU4, beta glucosidase 4 |
0.652 |
0.014 |
| 74 |
265057_at |
AT1G52140
|
unknown |
0.651 |
-0.030 |
| 75 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.650 |
-0.031 |
| 76 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
0.647 |
-0.043 |
| 77 |
254392_at |
AT4G21600
|
ENDO5, endonuclease 5 |
0.645 |
0.046 |
| 78 |
259912_at |
AT1G72670
|
iqd8, IQ-domain 8 |
0.640 |
0.023 |
| 79 |
263281_at |
AT2G14160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.639 |
-0.061 |
| 80 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
0.633 |
0.012 |
| 81 |
257165_at |
AT3G24330
|
[O-Glycosyl hydrolases family 17 protein] |
0.624 |
0.009 |
| 82 |
261903_at |
AT1G80740
|
CMT1, chromomethylase 1, DMT4, DNA METHYLTRANSFERASE 4 |
0.624 |
0.087 |
| 83 |
263365_at |
AT2G20550
|
[HSP40/DnaJ peptide-binding protein] |
0.624 |
0.053 |
| 84 |
257847_at |
AT3G13020
|
[hAT transposon superfamily protein] |
0.620 |
0.039 |
| 85 |
250143_at |
AT5G14670
|
ARFA1B, ADP-ribosylation factor A1B, ATARFA1B, ADP-ribosylation factor A1B |
0.611 |
0.059 |
| 86 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
0.602 |
-0.009 |
| 87 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
0.602 |
-0.058 |
| 88 |
259643_at |
AT1G68890
|
PHYLLO, PHYLLO |
0.591 |
-0.101 |
| 89 |
248734_at |
AT5G48090
|
ELP1, EDM2-like protein1 |
0.589 |
-0.000 |
| 90 |
258663_at |
AT3G08670
|
unknown |
0.586 |
-0.225 |
| 91 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
0.584 |
0.023 |
| 92 |
261382_at |
AT1G05470
|
CVP2, COTYLEDON VASCULAR PATTERN 2 |
0.582 |
-0.033 |
| 93 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.579 |
0.038 |
| 94 |
263609_at |
AT2G16250
|
[Leucine-rich repeat protein kinase family protein] |
0.579 |
-0.017 |
| 95 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
0.579 |
-0.089 |
| 96 |
264625_at |
AT1G09020
|
ATSNF4, SNF4, homolog of yeast sucrose nonfermenting 4 |
0.579 |
-0.052 |
| 97 |
251700_at |
AT3G56640
|
SEC15A, exocyst complex component sec15A |
0.578 |
-0.053 |
| 98 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.577 |
-0.137 |
| 99 |
264853_at |
AT2G17260
|
ATGLR2, ATGLR3.1, GLR2, glutamate receptor 2, GLR3.1 |
0.574 |
-0.026 |
| 100 |
265425_at |
AT2G20770
|
GCL2, GCR2-like 2 |
0.574 |
-0.054 |
| 101 |
261764_at |
AT1G15530
|
LecRK-S.1, L-type lectin receptor kinase S.1 |
0.574 |
-0.015 |
| 102 |
255417_at |
AT4G03190
|
AFB1, AUXIN SIGNALING F BOX PROTEIN 1, ATGRH1, GRH1, GRR1-like protein 1 |
0.573 |
-0.108 |
| 103 |
266314_at |
AT2G27040
|
AGO4, ARGONAUTE 4, OCP11, OVEREXPRESSOR OF CATIONIC PEROXIDASE 11 |
0.572 |
-0.049 |
| 104 |
247236_at |
AT5G64590
|
unknown |
0.568 |
0.041 |
| 105 |
245141_at |
AT2G45400
|
BEN1, BRI1-5 ENHANCED 1 |
0.568 |
0.032 |
| 106 |
249261_at |
AT5G41710
|
[Mutator-like transposase family, has a 6.2e-18 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.567 |
-0.034 |
| 107 |
256832_at |
AT3G22880
|
ARLIM15, ARABIDOPSIS HOMOLOG OF LILY MESSAGES INDUCED AT MEIOSIS 15, ATDMC1, ARABIDOPSIS THALIANA DISRUPTION OF MEIOTIC CONTROL 1, DMC1, DISRUPTION OF MEIOTIC CONTROL 1 |
0.563 |
0.053 |
| 108 |
263178_at |
AT1G05550
|
unknown |
0.558 |
-0.040 |
| 109 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.557 |
0.062 |
| 110 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.555 |
0.008 |
| 111 |
258477_at |
AT3G02680
|
ATNBS1, NBS1, nijmegen breakage syndrome 1 |
0.554 |
0.011 |
| 112 |
260210_at |
AT1G74420
|
ATFUT3, FUT3, fucosyltransferase 3 |
0.553 |
-0.089 |
| 113 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.551 |
0.008 |
| 114 |
245204_at |
AT5G12270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.550 |
-0.037 |
| 115 |
260493_at |
AT2G41830
|
[Uncharacterized protein] |
0.548 |
-0.049 |
| 116 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.545 |
0.019 |
| 117 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.545 |
0.152 |
| 118 |
261588_at |
AT1G01670
|
[RING/U-box superfamily protein] |
0.544 |
0.015 |
| 119 |
245560_at |
AT4G15480
|
UGT84A1 |
0.542 |
-0.006 |
| 120 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
0.541 |
0.082 |
| 121 |
252701_at |
AT3G43700
|
ATBPM6, BTB-POZ AND MATH DOMAIN 6, BPM6, BTB-POZ and MATH domain 6 |
0.539 |
0.050 |
| 122 |
260151_at |
AT1G52910
|
unknown |
0.539 |
0.031 |
| 123 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
0.538 |
-0.029 |
| 124 |
264990_at |
AT1G27210
|
[ARM repeat superfamily protein] |
0.538 |
-0.252 |
| 125 |
257275_at |
AT3G14450
|
CID9, CTC-interacting domain 9 |
0.537 |
0.103 |
| 126 |
249090_at |
AT5G43745
|
unknown |
0.535 |
-0.074 |
| 127 |
259263_at |
AT3G01010
|
[UDP-glucose/GDP-mannose dehydrogenase family protein] |
0.534 |
-0.045 |
| 128 |
249633_at |
AT5G37180
|
ATSUS5, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 5, SUS5, sucrose synthase 5 |
0.534 |
-0.018 |
| 129 |
261129_at |
AT1G04820
|
TOR2, TORTIFOLIA 2, TUA4, tubulin alpha-4 chain |
0.530 |
-0.022 |
| 130 |
253802_at |
AT4G28180
|
unknown |
0.526 |
-0.202 |
| 131 |
249397_at |
AT5G40230
|
UMAMIT37, Usually multiple acids move in and out Transporters 37 |
0.523 |
0.052 |
| 132 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
0.521 |
0.031 |
| 133 |
256492_at |
AT1G31490
|
[HXXXD-type acyl-transferase family protein] |
0.519 |
0.002 |
| 134 |
266715_at |
AT2G46780
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.519 |
-0.018 |
| 135 |
266718_at |
AT2G46800
|
ATCDF1, A. THALIANA CATION DIFFUSION FACILITATOR 1, ATMTP1, MTP1, metal tolerance protein 1, OZS1, OVERLY ZINC SENSITIVE 1, ZAT, zinc transporter of Arabidopsis thaliana, ZAT1, ZINC TRANSPORTER OF ARABIDOPSIS THALIANA 1 |
0.516 |
0.007 |
| 136 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.515 |
-0.030 |
| 137 |
255920_at |
AT5G28590
|
[DNA-binding family protein] |
0.514 |
-0.090 |
| 138 |
255258_at |
AT4G05060
|
[PapD-like superfamily protein] |
0.513 |
-0.056 |
| 139 |
258445_at |
AT3G22400
|
ATLOX5, Arabidopsis thaliana lipoxygenase 5, LOX5 |
0.513 |
-0.084 |
| 140 |
251010_at |
AT5G02550
|
unknown |
0.509 |
-0.128 |
| 141 |
255576_at |
AT4G01440
|
UMAMIT31, Usually multiple acids move in and out Transporters 31 |
0.506 |
0.032 |
| 142 |
257946_at |
AT3G21710
|
unknown |
0.506 |
0.022 |
| 143 |
259608_at |
AT1G27960
|
ECT9, evolutionarily conserved C-terminal region 9 |
0.503 |
0.048 |
| 144 |
265005_at |
AT1G61667
|
unknown |
0.503 |
-0.002 |
| 145 |
254140_at |
AT4G24610
|
unknown |
0.503 |
-0.019 |
| 146 |
252106_at |
AT3G51480
|
ATGLR3.6, glutamate receptor 3.6, GLR3.6, GLR3.6, glutamate receptor 3.6 |
0.502 |
-0.008 |
| 147 |
261921_at |
AT1G65900
|
unknown |
0.499 |
-0.003 |
| 148 |
251521_at |
AT3G59420
|
ACR4, crinkly4, CR4, crinkly4 |
0.498 |
-0.017 |
| 149 |
254349_at |
AT4G22250
|
[RING/U-box superfamily protein] |
0.497 |
0.029 |
| 150 |
246077_at |
AT5G20420
|
CHR42, chromatin remodeling 42 |
0.494 |
-0.031 |
| 151 |
251292_at |
AT3G61920
|
unknown |
0.492 |
-0.044 |
| 152 |
248435_at |
AT5G51210
|
OLEO3, oleosin3 |
0.491 |
0.012 |
| 153 |
254200_at |
AT4G24110
|
unknown |
0.485 |
0.137 |
| 154 |
253887_at |
AT4G27730
|
ATOPT6, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 6, OPT6, oligopeptide transporter 1 |
0.484 |
-0.077 |
| 155 |
265382_at |
AT2G16790
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.482 |
0.053 |
| 156 |
246915_at |
AT5G25880
|
ATNADP-ME3, Arabidopsis thaliana NADP-malic enzyme 3, NADP-ME3, NADP-malic enzyme 3 |
0.481 |
-0.019 |
| 157 |
265689_at |
AT2G24310
|
unknown |
0.480 |
-0.052 |
| 158 |
258122_at |
AT3G14570
|
ATGSL04, glucan synthase-like 4, atgsl4, gsl04, GSL04, glucan synthase-like 4 |
0.479 |
-0.038 |
| 159 |
255536_at |
AT4G01575
|
[serine protease inhibitor, Kazal-type family protein] |
0.479 |
-0.031 |
| 160 |
253050_at |
AT4G37450
|
AGP18, arabinogalactan protein 18, ATAGP18 |
0.477 |
-0.015 |
| 161 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.477 |
-0.022 |
| 162 |
258951_at |
AT3G01380
|
[transferases] |
0.476 |
-0.087 |
| 163 |
262920_at |
AT1G79500
|
AtkdsA1 |
0.476 |
0.028 |
| 164 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.476 |
0.013 |
| 165 |
247397_at |
AT5G62940
|
DOF5.6, DNA BINDING WITH ONE FINGER 5.6, HCA2, HIGH CAMBIAL ACTIVITY2 |
0.475 |
-0.002 |
| 166 |
267266_at |
AT2G23150
|
ATNRAMP3, NRAMP3, natural resistance-associated macrophage protein 3 |
0.472 |
-0.002 |
| 167 |
247195_at |
AT5G65500
|
[U-box domain-containing protein kinase family protein] |
0.472 |
0.020 |
| 168 |
245859_at |
AT5G28290
|
ATNEK3, NIMA-related kinase 3, NEK3, NIMA-related kinase 3 |
0.472 |
-0.115 |
| 169 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.471 |
-0.034 |
| 170 |
263363_at |
AT2G03850
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.466 |
0.054 |
| 171 |
248247_at |
AT5G53210
|
SPCH, SPEECHLESS |
0.463 |
0.024 |
| 172 |
264712_at |
AT1G09810
|
ECT11, evolutionarily conserved C-terminal region 11 |
0.463 |
-0.024 |
| 173 |
261551_at |
AT1G63440
|
HMA5, heavy metal atpase 5 |
0.462 |
-0.014 |
| 174 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.456 |
0.028 |
| 175 |
257342_at |
AT1G42365
|
[gypsy-like retrotransposon family (Athila), has a 9.0e-234 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.456 |
0.025 |
| 176 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.456 |
0.008 |
| 177 |
265559_at |
AT2G05530
|
[Glycine-rich protein family] |
0.456 |
0.111 |
| 178 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.454 |
-0.021 |
| 179 |
260925_at |
AT1G21340
|
[Dof-type zinc finger DNA-binding family protein] |
0.453 |
-0.024 |
| 180 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
0.453 |
-0.028 |
| 181 |
263910_at |
AT2G36550
|
unknown |
0.451 |
-0.026 |
| 182 |
264916_at |
AT1G60810
|
ACLA-2, ATP-citrate lyase A-2 |
0.451 |
0.044 |
| 183 |
246329_at |
AT3G43610
|
[Spc97 / Spc98 family of spindle pole body (SBP) component] |
0.449 |
-0.082 |
| 184 |
246063_at |
AT5G19340
|
unknown |
0.449 |
-0.087 |
| 185 |
246512_at |
AT5G15630
|
COBL4, COBRA-LIKE4, IRX6, IRREGULAR XYLEM 6 |
0.448 |
0.024 |
| 186 |
263047_at |
AT2G17630
|
PSAT2, phosphoserine aminotransferase 2 |
0.448 |
0.022 |
| 187 |
266917_at |
AT2G45830
|
DTA2, downstream target of AGL15 2 |
0.448 |
0.002 |
| 188 |
252769_at |
AT3G42850
|
[Mevalonate/galactokinase family protein] |
0.447 |
0.081 |
| 189 |
259641_at |
AT1G69020
|
[Prolyl oligopeptidase family protein] |
0.447 |
-0.007 |
| 190 |
257422_at |
AT1G11940
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.447 |
0.039 |
| 191 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-1.623 |
0.133 |
| 192 |
249482_at |
AT5G38980
|
unknown |
-1.606 |
-0.022 |
| 193 |
252462_at |
AT3G47250
|
unknown |
-1.448 |
0.061 |
| 194 |
265611_at |
AT2G25510
|
unknown |
-1.443 |
-0.020 |
| 195 |
256442_at |
AT3G10930
|
unknown |
-1.415 |
0.267 |
| 196 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.414 |
0.028 |
| 197 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-1.389 |
0.061 |
| 198 |
252346_at |
AT3G48650
|
[pseudogene] |
-1.330 |
0.050 |
| 199 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-1.310 |
0.091 |
| 200 |
262442_at |
AT1G47420
|
SDH5, succinate dehydrogenase 5 |
-1.256 |
0.022 |
| 201 |
248415_at |
AT5G51620
|
[Uncharacterised protein family (UPF0172)] |
-1.239 |
0.108 |
| 202 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-1.229 |
0.069 |
| 203 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-1.229 |
0.162 |
| 204 |
253422_at |
AT4G32240
|
unknown |
-1.228 |
0.067 |
| 205 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-1.220 |
0.001 |
| 206 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
-1.211 |
0.010 |
| 207 |
262958_at |
AT1G54410
|
AtHIRD11, HIRD11, dehydrin 11kDa |
-1.193 |
0.031 |
| 208 |
258114_at |
AT3G14660
|
CYP72A13, cytochrome P450, family 72, subfamily A, polypeptide 13 |
-1.174 |
0.034 |
| 209 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
-1.168 |
0.051 |
| 210 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.129 |
0.019 |
| 211 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-1.113 |
0.086 |
| 212 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-1.108 |
0.010 |
| 213 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-1.104 |
-0.066 |
| 214 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
-1.103 |
0.114 |
| 215 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
-1.089 |
0.049 |
| 216 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-1.080 |
0.255 |
| 217 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.074 |
0.093 |
| 218 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
-1.066 |
-0.016 |
| 219 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
-1.061 |
0.026 |
| 220 |
253096_at |
AT4G37330
|
CYP81D4, cytochrome P450, family 81, subfamily D, polypeptide 4 |
-1.061 |
-0.016 |
| 221 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-1.059 |
0.229 |
| 222 |
254910_at |
AT4G11175
|
[Nucleic acid-binding, OB-fold-like protein] |
-1.056 |
0.082 |
| 223 |
262638_at |
AT1G06650
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.050 |
-0.021 |
| 224 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.046 |
0.051 |
| 225 |
267127_at |
AT2G23610
|
ATMES3, ARABIDOPSIS THALIANA METHYL ESTERASE 3, MES3, methyl esterase 3 |
-1.044 |
0.024 |
| 226 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
-1.039 |
0.036 |
| 227 |
246623_at |
AT1G48920
|
ATNUC-L1, nucleolin like 1, NUC-L1, nucleolin like 1, PARL1, PARALLEL 1 |
-1.038 |
0.037 |
| 228 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-1.032 |
-0.023 |
| 229 |
258027_at |
AT3G19515
|
unknown |
-1.032 |
0.019 |
| 230 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
-1.023 |
-0.060 |
| 231 |
256568_at |
AT3G19520
|
unknown |
-1.019 |
0.068 |
| 232 |
252661_at |
AT3G44450
|
unknown |
-1.016 |
0.162 |
| 233 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-1.015 |
0.056 |
| 234 |
247722_at |
AT5G59150
|
ATRAB-A2D, ARABIDOPSIS RAB GTPASE HOMOLOG A2D, ATRABA2D, RAB GTPase homolog A2D, RABA2D, RAB GTPase homolog A2D |
-1.010 |
0.049 |
| 235 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.009 |
0.060 |
| 236 |
256603_at |
AT3G28270
|
unknown |
-1.003 |
0.068 |
| 237 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.997 |
-0.022 |
| 238 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.989 |
-0.091 |
| 239 |
263182_at |
AT1G05575
|
unknown |
-0.982 |
0.216 |
| 240 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.977 |
0.048 |
| 241 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.974 |
0.040 |
| 242 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
-0.973 |
0.116 |
| 243 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
-0.970 |
0.014 |
| 244 |
249029_at |
AT5G44870
|
LAZ5, LAZARUS 5, TTR1, tolerance to Tobacco ringspot virus 1 |
-0.958 |
0.015 |
| 245 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.954 |
0.088 |
| 246 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.948 |
0.052 |
| 247 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
-0.946 |
0.006 |
| 248 |
260005_at |
AT1G67920
|
unknown |
-0.943 |
0.033 |
| 249 |
248967_at |
AT5G45350
|
[proline-rich family protein] |
-0.934 |
-0.008 |
| 250 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.922 |
-0.033 |
| 251 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
-0.913 |
0.138 |
| 252 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
-0.907 |
-0.019 |
| 253 |
261221_at |
AT1G19960
|
unknown |
-0.906 |
-0.065 |
| 254 |
255504_at |
AT4G02200
|
[Drought-responsive family protein] |
-0.906 |
0.125 |
| 255 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
-0.903 |
0.055 |
| 256 |
266480_at |
AT2G31130
|
unknown |
-0.898 |
0.065 |
| 257 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-0.894 |
0.096 |
| 258 |
260031_at |
AT1G68790
|
CRWN3, CROWDED NUCLEI 3, LINC3, LITTLE NUCLEI3 |
-0.894 |
-0.062 |
| 259 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.892 |
0.023 |
| 260 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
-0.890 |
0.074 |
| 261 |
264843_at |
AT1G03400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.883 |
0.035 |
| 262 |
253918_at |
AT4G27320
|
ATPHOS34, PHOS34 |
-0.877 |
-0.029 |
| 263 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
-0.872 |
0.093 |
| 264 |
246476_at |
AT5G16730
|
[LOCATED IN: chloroplast] |
-0.869 |
0.027 |
| 265 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.868 |
0.042 |
| 266 |
246421_at |
AT5G16880
|
[Target of Myb protein 1] |
-0.864 |
0.045 |
| 267 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.860 |
0.133 |
| 268 |
262615_at |
AT1G13950
|
ATELF5A-1, EUKARYOTIC ELONGATION FACTOR 5A-1, EIF-5A, EIF5A, EUKARYOTIC ELONGATION FACTOR 5A, ELF5A-1, eukaryotic elongation factor 5A-1 |
-0.855 |
-0.076 |
| 269 |
249625_at |
AT5G37480
|
unknown |
-0.855 |
0.095 |
| 270 |
258982_at |
AT3G08870
|
LecRK-VI.1, L-type lectin receptor kinase VI.1 |
-0.853 |
-0.056 |
| 271 |
248972_at |
AT5G45010
|
ATDSS1(V), DSS1 homolog on chromosome V, DSS1(V), DSS1 homolog on chromosome V |
-0.849 |
0.053 |
| 272 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
-0.848 |
0.171 |
| 273 |
252294_at |
AT3G49010
|
ATBBC1, breast basic conserved 1, BBC1, breast basic conserved 1, RSU2, 40S RIBOSOMAL PROTEIN |
-0.844 |
0.005 |
| 274 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.842 |
0.016 |
| 275 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
-0.839 |
0.058 |
| 276 |
252345_at |
AT3G48640
|
unknown |
-0.837 |
-0.033 |
| 277 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.837 |
0.102 |
| 278 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.837 |
-0.023 |
| 279 |
256796_at |
AT3G22210
|
unknown |
-0.834 |
0.096 |
| 280 |
261317_at |
AT1G53030
|
[Cytochrome C oxidase copper chaperone (COX17)] |
-0.833 |
0.090 |
| 281 |
249165_at |
AT5G42810
|
ATIPK1, inositol-pentakisphosphate 2-kinase 1, IPK1, inositol-pentakisphosphate 2-kinase 1 |
-0.828 |
0.097 |
| 282 |
263192_at |
AT1G36160
|
ACC1, acetyl-CoA carboxylase 1, AT-ACC1, EMB22, EMBRYO DEFECTIVE 22, GK, GURKE, GSD1, GLOSSYHEAD 1, PAS3, PASTICCINO 3, SFR3, SENSITIVE TO FREEZING 3 |
-0.827 |
-0.030 |
| 283 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
-0.825 |
-0.009 |
| 284 |
259102_at |
AT3G11660
|
NHL1, NDR1/HIN1-like 1 |
-0.824 |
-0.026 |
| 285 |
254278_at |
AT4G22740
|
[glycine-rich protein] |
-0.820 |
0.111 |
| 286 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
-0.818 |
0.124 |
| 287 |
245566_at |
AT4G14610
|
[pseudogene] |
-0.815 |
-0.003 |
| 288 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
-0.808 |
-0.028 |
| 289 |
255793_at |
AT2G33250
|
unknown |
-0.805 |
0.058 |
| 290 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.803 |
0.095 |
| 291 |
254190_at |
AT4G23885
|
unknown |
-0.802 |
0.056 |
| 292 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.798 |
0.019 |
| 293 |
260155_at |
AT1G52870
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.796 |
0.047 |
| 294 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
-0.792 |
0.064 |
| 295 |
267374_at |
AT2G26230
|
[uricase / urate oxidase / nodulin 35, putative] |
-0.790 |
0.014 |
| 296 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.790 |
0.074 |
| 297 |
263875_at |
AT2G21970
|
SEP2, stress enhanced protein 2 |
-0.788 |
0.116 |
| 298 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.785 |
0.034 |
| 299 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.785 |
0.026 |
| 300 |
258092_at |
AT3G14595
|
[Ribosomal protein L18ae family] |
-0.783 |
0.044 |
| 301 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.781 |
0.036 |
| 302 |
249839_at |
AT5G23405
|
[HMG-box (high mobility group) DNA-binding family protein] |
-0.780 |
0.066 |
| 303 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.780 |
0.150 |
| 304 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.778 |
0.069 |
| 305 |
247999_at |
AT5G56150
|
UBC30, ubiquitin-conjugating enzyme 30 |
-0.773 |
0.018 |
| 306 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.768 |
-0.028 |
| 307 |
266482_at |
AT2G47640
|
[Small nuclear ribonucleoprotein family protein] |
-0.767 |
0.050 |
| 308 |
265909_at |
AT2G25720
|
unknown |
-0.766 |
0.096 |
| 309 |
260870_at |
AT1G43890
|
ATRAB-C1, ARABIDOPSIS RAB GTPASE HOMOLOG C1, ATRAB18, ARABIDOPSIS RAB GTPASE HOMOLOG B18, ATRABC1, ARABIDOPSIS RAB GTPASE HOMOLOG C1, RAB18, RAB GTPASE HOMOLOG B18, RAB18-1, RAB GTPASE HOMOLOG 18-1, RABC1, RAB GTPASE HOMOLOG C1 |
-0.762 |
0.025 |
| 310 |
254481_at |
AT4G20480
|
[Putative endonuclease or glycosyl hydrolase] |
-0.762 |
-0.036 |
| 311 |
262669_at |
AT1G62850
|
[Class I peptide chain release factor] |
-0.759 |
0.054 |
| 312 |
245924_at |
AT5G28750
|
[Bacterial sec-independent translocation protein mttA/Hcf106] |
-0.759 |
0.051 |
| 313 |
255923_at |
AT1G22180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.754 |
0.120 |
| 314 |
248329_at |
AT5G52780
|
unknown |
-0.750 |
0.096 |
| 315 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-0.745 |
0.199 |
| 316 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.745 |
0.017 |
| 317 |
260211_at |
AT1G74440
|
unknown |
-0.744 |
0.033 |
| 318 |
245451_at |
AT4G16890
|
BAL, BALL, SNC1, SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 |
-0.742 |
0.031 |
| 319 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.737 |
-0.081 |
| 320 |
262175_at |
AT1G74880
|
NDH-O, NAD(P)H:plastoquinone dehydrogenase complex subunit O, NdhO, NADH dehydrogenase-like complex ) |
-0.737 |
-0.038 |
| 321 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.734 |
0.034 |
| 322 |
253990_at |
AT4G26160
|
ACHT1, atypical CYS HIS rich thioredoxin 1 |
-0.728 |
0.016 |
| 323 |
266310_at |
AT2G26990
|
ATCSN2, COP12, CONSTITUTIVE PHOTOMORPHOGENIC 12, CSN2, FUS12, FUSCA 12 |
-0.728 |
0.054 |
| 324 |
254955_at |
AT4G10920
|
KELP |
-0.727 |
0.043 |
| 325 |
251542_at |
AT3G58760
|
[Integrin-linked protein kinase family] |
-0.724 |
0.022 |
| 326 |
262631_at |
AT1G06500
|
unknown |
-0.724 |
0.076 |
| 327 |
256216_at |
AT1G56340
|
AtCRT1a, CRT1, calreticulin 1, CRT1a, calreticulin 1a |
-0.722 |
-0.010 |
| 328 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.721 |
-0.068 |
| 329 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
-0.720 |
0.043 |
| 330 |
245384_at |
AT4G16790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.720 |
-0.254 |
| 331 |
257771_at |
AT3G23000
|
ATSR2, ATSRPK1, CIPK7, CBL-interacting protein kinase 7, PKS7, SnRK3.10, SNF1-RELATED PROTEIN KINASE 3.10 |
-0.718 |
0.076 |
| 332 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
-0.718 |
-0.022 |
| 333 |
250077_at |
AT5G16680
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.713 |
0.007 |
| 334 |
249886_at |
AT5G22320
|
[Leucine-rich repeat (LRR) family protein] |
-0.712 |
0.051 |
| 335 |
253545_at |
AT4G31310
|
[AIG2-like (avirulence induced gene) family protein] |
-0.711 |
0.089 |
| 336 |
251670_at |
AT3G57190
|
PrfB3, peptide chain release factor 3 |
-0.709 |
0.027 |
| 337 |
249161_at |
AT5G42790
|
ARS5, ARSENIC TOLERANCE 5, ATPSM30, PAF1, proteasome alpha subunit F1 |
-0.707 |
0.057 |
| 338 |
248146_at |
AT5G54940
|
[Translation initiation factor SUI1 family protein] |
-0.707 |
0.066 |
| 339 |
261270_at |
AT1G26630
|
ATELF5A-2, EUKARYOTIC ELONGATION FACTOR 5A-2, ELF5A-2, EUKARYOTIC ELONGATION FACTOR 5A-2, FBR12, FUMONISIN B1-RESISTANT12 |
-0.706 |
-0.006 |
| 340 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.701 |
0.040 |
| 341 |
246791_at |
AT5G27280
|
[Zim17-type zinc finger protein] |
-0.701 |
0.060 |
| 342 |
250800_at |
AT5G05370
|
[Cytochrome b-c1 complex, subunit 8 protein] |
-0.700 |
0.052 |
| 343 |
256744_at |
AT3G29350
|
AHP2, histidine-containing phosphotransmitter 2 |
-0.700 |
0.031 |
| 344 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.699 |
0.048 |
| 345 |
250395_at |
AT5G10950
|
[Tudor/PWWP/MBT superfamily protein] |
-0.698 |
0.068 |
| 346 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.696 |
-0.042 |
| 347 |
261607_at |
AT1G49660
|
AtCXE5, carboxyesterase 5, CXE5, carboxyesterase 5 |
-0.695 |
0.144 |
| 348 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
-0.695 |
0.024 |
| 349 |
246144_at |
AT5G20110
|
[Dynein light chain type 1 family protein] |
-0.694 |
-0.067 |
| 350 |
264673_at |
AT1G09795
|
ATATP-PRT2, ATP phosphoribosyl transferase 2, ATP-PRT2, ATP phosphoribosyl transferase 2, HISN1B |
-0.692 |
-0.028 |
| 351 |
260416_at |
AT1G69670
|
ATCUL3B, ARABIDOPSIS THALIANA CULLIN 3B, CUL3B, cullin 3B |
-0.692 |
0.036 |
| 352 |
256617_at |
AT3G22240
|
unknown |
-0.684 |
0.025 |
| 353 |
249393_at |
AT5G40170
|
AtRLP54, receptor like protein 54, RLP54, receptor like protein 54 |
-0.684 |
-0.042 |
| 354 |
253174_at |
AT4G35090
|
CAT2, catalase 2 |
-0.683 |
-0.005 |
| 355 |
245045_at |
AT2G26590
|
RPN13, regulatory particle non-ATPase 13 |
-0.682 |
0.027 |
| 356 |
247488_at |
AT5G61820
|
unknown |
-0.681 |
0.047 |
| 357 |
261145_at |
AT1G19730
|
ATH4, thioredoxin H-type 4, ATTRX4 |
-0.681 |
0.064 |
| 358 |
245114_at |
AT2G41630
|
AtTFIIB1, TFIIB, transcription factor IIB, TFIIB1, transcription factor IIB 1 |
-0.679 |
0.047 |
| 359 |
265245_at |
AT2G43060
|
AtIBH1, IBH1, ILI1 binding bHLH 1 |
-0.677 |
-0.041 |
| 360 |
247373_at |
AT5G63150
|
unknown |
-0.676 |
0.059 |
| 361 |
251281_at |
AT3G61640
|
AGP20, arabinogalactan protein 20, AtAGP20, Arabinogalactan protein 20 |
-0.675 |
-0.175 |
| 362 |
255654_at |
AT4G00970
|
CRK41, cysteine-rich RLK (RECEPTOR-like protein kinase) 41 |
-0.673 |
0.023 |
| 363 |
249176_at |
AT5G42980
|
ATH3, thioredoxin H-type 3, ATTRX3, thioredoxin 3, ATTRXH3, TRX3, thioredoxin 3, TRXH3, THIOREDOXIN H3 |
-0.672 |
0.010 |
| 364 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
-0.671 |
-0.020 |
| 365 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
-0.668 |
0.038 |
| 366 |
246921_at |
AT5G25080
|
[Sas10/Utp3/C1D family] |
-0.668 |
0.014 |
| 367 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
-0.667 |
0.058 |
| 368 |
260128_at |
AT1G36310
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.666 |
0.005 |
| 369 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-0.666 |
0.003 |
| 370 |
257249_at |
AT3G24180
|
[Beta-glucosidase, GBA2 type family protein] |
-0.665 |
-0.020 |
| 371 |
260089_at |
AT1G73170
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.665 |
0.070 |
| 372 |
246069_at |
AT5G20220
|
[zinc knuckle (CCHC-type) family protein] |
-0.663 |
0.001 |
| 373 |
261535_at |
AT1G01725
|
unknown |
-0.663 |
0.057 |
| 374 |
264699_at |
AT1G69980
|
unknown |
-0.662 |
0.073 |
| 375 |
252824_at |
AT4G40030
|
H3.3, histone 3.3 |
-0.657 |
0.006 |
| 376 |
255154_at |
AT4G08220
|
[Mutator-like transposase family, has a 5.3e-67 P-value blast match to Q9SUF8 /145-308 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-0.655 |
0.013 |
| 377 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.655 |
-0.078 |
| 378 |
254628_at |
AT4G18593
|
[dual specificity protein phosphatase-related] |
-0.655 |
0.031 |
| 379 |
257949_at |
AT3G21750
|
UGT71B1, UDP-glucosyl transferase 71B1 |
-0.654 |
0.011 |