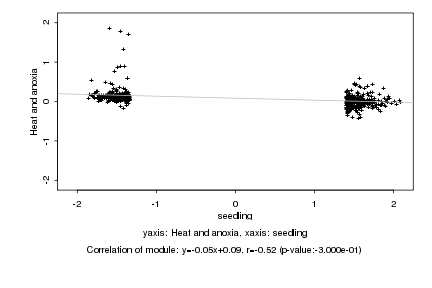
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
seedling |
Log2 signal ratio
Heat and anoxia |
| 1 |
265560_at |
AT2G05520
|
ATGRP-3, GLYCINE-RICH PROTEIN 3, ATGRP3, ARABIDOPSIS GLYCINE RICH PROTEIN 3, GRP-3, glycine-rich protein 3, GRP3, GLYCINE-RICH PROTEIN 3 |
2.082 |
-0.015 |
| 2 |
251762_at |
AT3G55800
|
SBPASE, sedoheptulose-bisphosphatase |
2.066 |
0.027 |
| 3 |
259612_at |
AT1G52300
|
[Zinc-binding ribosomal protein family protein] |
2.029 |
-0.068 |
| 4 |
245195_at |
AT1G67740
|
PSBY, photosystem II BY, YCF32 |
2.005 |
0.004 |
| 5 |
263764_at |
AT2G21410
|
VHA-A2, vacuolar proton ATPase A2 |
1.972 |
-0.039 |
| 6 |
265649_at |
AT2G27510
|
ATFD3, ferredoxin 3, FD3, ferredoxin 3 |
1.937 |
0.099 |
| 7 |
252130_at |
AT3G50820
|
OEC33, OXYGEN EVOLVING COMPLEX SUBUNIT 33 KDA, PSBO-2, PHOTOSYSTEM II SUBUNIT O-2, PSBO2, photosystem II subunit O-2 |
1.923 |
0.145 |
| 8 |
258433_at |
AT3G16640
|
TCTP, translationally controlled tumor protein |
1.921 |
0.055 |
| 9 |
257807_at |
AT3G26650
|
GAPA, glyceraldehyde 3-phosphate dehydrogenase A subunit, GAPA-1, GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT 1 |
1.916 |
-0.016 |
| 10 |
255457_at |
AT4G02770
|
PSAD-1, photosystem I subunit D-1 |
1.909 |
0.004 |
| 11 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
1.894 |
-0.116 |
| 12 |
256679_at |
AT3G52300
|
ATPQ, ATP synthase D chain, mitochondrial |
1.885 |
0.005 |
| 13 |
251185_at |
AT3G62870
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
1.878 |
-0.086 |
| 14 |
255629_at |
AT4G00860
|
AT0ZI1, Arabidopsis thaliana ozone-induced protein 1, ATOZI1 |
1.873 |
0.084 |
| 15 |
248582_at |
AT5G49910
|
cpHsc70-2, chloroplast heat shock protein 70-2, HSC70-7, HEAT SHOCK PROTEIN 70-7 |
1.870 |
0.335 |
| 16 |
263676_at |
AT1G09340
|
CRB, chloroplast RNA binding, CSP41B, CHLOROPLAST STEM-LOOP BINDING PROTEIN OF 41 KDA, HIP1.3, heteroglycan-interacting protein 1.3 |
1.857 |
-0.047 |
| 17 |
252973_s_at |
AT4G38740
|
ROC1, rotamase CYP 1 |
1.838 |
0.005 |
| 18 |
250898_at |
AT5G03300
|
ADK2, adenosine kinase 2 |
1.835 |
-0.084 |
| 19 |
257173_at |
AT3G23810
|
ATSAHH2, S-ADENOSYL-L-HOMOCYSTEINE (SAH) HYDROLASE 2, SAHH2, S-adenosyl-l-homocysteine (SAH) hydrolase 2 |
1.829 |
-0.250 |
| 20 |
246673_at |
AT5G30510
|
ARRPS1, PRPS1, plastid ribosomal protein S1, RPS1, ribosomal protein S1 |
1.824 |
-0.085 |
| 21 |
256964_at |
AT3G13520
|
AGP12, arabinogalactan protein 12, ATAGP12 |
1.821 |
0.193 |
| 22 |
252929_at |
AT4G38970
|
AtFBA2, FBA2, fructose-bisphosphate aldolase 2 |
1.808 |
-0.071 |
| 23 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
1.803 |
-0.201 |
| 24 |
257749_at |
AT3G18780
|
ACT2, actin 2, DER1, DEFORMED ROOT HAIRS 1, ENL2, ENHANCER OF LRX1 2, FIZ2, FRIZZY AND KINKED SHOOTS 2, LSR2, LIGHT STRESS-REGULATED 2 |
1.802 |
0.033 |
| 25 |
259112_at |
AT3G05560
|
[Ribosomal L22e protein family] |
1.779 |
-0.133 |
| 26 |
255482_at |
AT4G02510
|
ATTOC159, PPI2, PLASTID PROTEIN IMPORT 2, TOC159, translocon at the outer envelope membrane of chloroplasts 159, TOC160, TRANSLOCON AT THE OUTER ENVELOPE MEMBRANE OF CHLOROPLASTS 160, TOC86, TRANSLOCON AT THE OUTER ENVELOPE MEMBRANE OF CHLOROPLASTS 86 |
1.772 |
-0.069 |
| 27 |
257190_at |
AT3G13120
|
[Ribosomal protein S10p/S20e family protein] |
1.764 |
-0.100 |
| 28 |
265996_at |
AT2G24200
|
LAP1, leucyl aminopeptidase 1 |
1.757 |
0.034 |
| 29 |
261412_at |
AT1G07890
|
APX1, ascorbate peroxidase 1, ATAPX01, ATAPX1, CS1, MEE6, maternal effect embryo arrest 6 |
1.753 |
0.096 |
| 30 |
252880_at |
AT4G39730
|
PLAT1, PLAT domain protein 1 |
1.749 |
-0.011 |
| 31 |
251845_at |
AT3G54540
|
ABCF4, ATP-binding cassette F4, ATGCN4, general control non-repressible 4, GCN4, general control non-repressible 4 |
1.749 |
-0.000 |
| 32 |
252409_at |
AT3G47650
|
[DnaJ/Hsp40 cysteine-rich domain superfamily protein] |
1.744 |
-0.129 |
| 33 |
256835_at |
AT3G22890
|
APS1, ATP sulfurylase 1 |
1.732 |
0.021 |
| 34 |
254981_at |
AT4G10480
|
[Nascent polypeptide-associated complex (NAC), alpha subunit family protein] |
1.729 |
-0.120 |
| 35 |
252971_at |
AT4G38770
|
ATPRP4, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 4, PRP4, proline-rich protein 4 |
1.729 |
-0.164 |
| 36 |
252235_at |
AT3G49910
|
[Translation protein SH3-like family protein] |
1.728 |
-0.040 |
| 37 |
255435_at |
AT4G03280
|
PETC, photosynthetic electron transfer C, PGR1, PROTON GRADIENT REGULATION 1 |
1.727 |
0.008 |
| 38 |
263483_at |
AT2G04030
|
AtHsp90.5, HEAT SHOCK PROTEIN 90.5, AtHsp90C, CR88, EMB1956, EMBRYO DEFECTIVE 1956, Hsp88.1, HEAT SHOCK PROTEIN 88.1, HSP90.5, HEAT SHOCK PROTEIN 90.5 |
1.726 |
0.237 |
| 39 |
254480_at |
AT4G20360
|
ATRAB8D, ATRABE1B, RAB GTPase homolog E1B, RABE1b, RAB GTPase homolog E1B |
1.725 |
-0.082 |
| 40 |
255456_at |
AT4G02920
|
unknown |
1.721 |
0.456 |
| 41 |
255442_at |
AT4G02580
|
[NADH-ubiquinone oxidoreductase 24 kDa subunit, putative] |
1.719 |
0.033 |
| 42 |
264595_at |
AT1G04750
|
AT VAMP7B, ATVAMP721, VAMP721, vesicle-associated membrane protein 721, VAMP7B, VESICLE-ASSOCIATED MEMBRANE PROTEIN 7B |
1.712 |
0.129 |
| 43 |
262468_at |
AT1G50200
|
ACD, ALATS, Alanyl-tRNA synthetase |
1.711 |
0.058 |
| 44 |
262884_at |
AT1G64720
|
CP5 |
1.707 |
0.018 |
| 45 |
248825_at |
AT5G47030
|
[ATPase, F1 complex, delta/epsilon subunit] |
1.706 |
-0.022 |
| 46 |
256035_at |
AT1G07140
|
SIRANBP |
1.705 |
0.004 |
| 47 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
1.703 |
0.234 |
| 48 |
249700_at |
AT5G35530
|
[Ribosomal protein S3 family protein] |
1.703 |
-0.003 |
| 49 |
261119_at |
AT1G75350
|
emb2184, embryo defective 2184 |
1.698 |
0.012 |
| 50 |
255104_at |
AT4G08685
|
SAH7 |
1.693 |
-0.046 |
| 51 |
262341_at |
AT1G64230
|
UBC28, ubiquitin-conjugating enzyme 28 |
1.692 |
0.059 |
| 52 |
249796_at |
AT5G23540
|
[Mov34/MPN/PAD-1 family protein] |
1.691 |
0.033 |
| 53 |
262980_at |
AT1G75680
|
AtGH9B7, glycosyl hydrolase 9B7, GH9B7, glycosyl hydrolase 9B7 |
1.691 |
0.001 |
| 54 |
260165_at |
AT1G79850
|
CS17, PDE347, PIGMENT DEFECTIVE 347, PRPS17, PLASTID RIBOSOMAL SMALL SUBUNIT PROTEIN 17, RPS17, ribosomal protein S17 |
1.686 |
-0.075 |
| 55 |
255691_at |
AT4G00370
|
ANTR2, PHT4;4, anion transporter 2 |
1.681 |
-0.147 |
| 56 |
258677_at |
AT3G08730
|
ATPK1, ARABIDOPSIS THALIANA PROTEIN-SERINE KINASE 1, ATPK6, ARABIDOPSIS THALIANA PROTEIN-SERINE KINASE 6, ATS6K1, PK1, protein-serine kinase 1, PK6, ROTEIN-SERINE KINASE 6, S6K1, P70 RIBOSOMAL S6 KINASE |
1.676 |
0.158 |
| 57 |
245686_at |
AT5G22060
|
ATJ2, ARABIDOPSIS THALIANA DNAJ HOMOLOGUE 2, J2, DNAJ homologue 2 |
1.668 |
0.394 |
| 58 |
248950_at |
AT5G45390
|
CLPP4, CLP protease P4, NCLPP4, NUCLEAR-ENCODED CLP PROTEASE P4 |
1.667 |
-0.068 |
| 59 |
252955_at |
AT4G38630
|
ATMCB1, MBP1, MULTIUBIQUITIN CHAIN BINDING PROTEIN 1, MCB1, MULTIUBIQUITIN-CHAIN-BINDING PROTEIN 1, RPN10, regulatory particle non-ATPase 10 |
1.665 |
0.008 |
| 60 |
252670_at |
AT3G44110
|
ATJ, ATJ3, DNAJ homologue 3, J3 |
1.664 |
0.384 |
| 61 |
264445_at |
AT1G27290
|
unknown |
1.662 |
0.205 |
| 62 |
246735_at |
AT5G27670
|
HTA7, histone H2A 7 |
1.659 |
0.026 |
| 63 |
258285_at |
AT3G16140
|
PSAH-1, photosystem I subunit H-1 |
1.658 |
0.055 |
| 64 |
260718_at |
AT1G48110
|
ECT7, evolutionarily conserved C-terminal region 7 |
1.649 |
-0.020 |
| 65 |
252845_at |
AT3G42050
|
[vacuolar ATP synthase subunit H family protein] |
1.648 |
0.011 |
| 66 |
258709_at |
AT3G09500
|
[Ribosomal L29 family protein ] |
1.643 |
-0.005 |
| 67 |
261270_at |
AT1G26630
|
ATELF5A-2, EUKARYOTIC ELONGATION FACTOR 5A-2, ELF5A-2, EUKARYOTIC ELONGATION FACTOR 5A-2, FBR12, FUMONISIN B1-RESISTANT12 |
1.634 |
-0.007 |
| 68 |
267005_at |
AT2G34460
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.630 |
0.047 |
| 69 |
266461_at |
AT2G47730
|
ATGSTF5, GLUTATHIONE S-TRANSFERASE (CLASS PHI) 5, ATGSTF8, Arabidopsis thaliana glutathione S-transferase phi 8, GST6, GSTF8, glutathione S-transferase phi 8 |
1.630 |
0.343 |
| 70 |
259096_at |
AT3G04840
|
[Ribosomal protein S3Ae] |
1.629 |
-0.107 |
| 71 |
263802_at |
AT2G40430
|
unknown |
1.628 |
0.015 |
| 72 |
266716_at |
AT2G46820
|
CURT1B, CURVATURE THYLAKOID 1B, PSAP, PSI-P, photosystem I P subunit, PTAC8, PLASTID TRANSCRIPTIONALLY ACTIVE 8, TMP14, THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA |
1.622 |
-0.008 |
| 73 |
252974_at |
AT4G38800
|
ATMTAN1, ARABIDOPSIS METHYLTHIOADENOSINE NUCLEOSIDASE 1, ATMTN1, ARABIDOPSIS METHYLTHIOADENOSINE NUCLEOSIDASE 1, MTAN1, METHYLTHIOADENOSINE NUCLEOSIDASE 1, MTN1, methylthioadenosine nucleosidase 1 |
1.622 |
0.050 |
| 74 |
256267_at |
AT3G12260
|
[LYR family of Fe/S cluster biogenesis protein] |
1.619 |
0.005 |
| 75 |
265671_at |
AT2G32060
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
1.619 |
-0.068 |
| 76 |
247968_at |
AT5G56670
|
[Ribosomal protein S30 family protein] |
1.617 |
-0.086 |
| 77 |
261919_at |
AT1G65980
|
TPX1, thioredoxin-dependent peroxidase 1 |
1.614 |
0.185 |
| 78 |
259239_at |
AT3G11510
|
[Ribosomal protein S11 family protein] |
1.611 |
-0.095 |
| 79 |
252982_at |
AT4G38130
|
ATHD1, ARABIDOPSIS HISTONE DEACETYLASE 1, ATHDA19, ARABIDOPSIS HISTONE DEACETYLASE 19, HD1, histone deacetylase 1, HDA1, HDA19, HISTONE DEACETYLASE 19, RPD3A |
1.606 |
0.047 |
| 80 |
257033_at |
AT3G19170
|
ATPREP1, presequence protease 1, ATZNMP, PREP1, presequence protease 1 |
1.601 |
-0.050 |
| 81 |
266701_at |
AT2G19760
|
PFN1, PROFILIN 1, PRF1, profilin 1 |
1.599 |
0.066 |
| 82 |
261979_at |
AT1G37130
|
ATNR2, ARABIDOPSIS NITRATE REDUCTASE 2, B29, CHL3, CHLORATE RESISTANT 3, NIA2, nitrate reductase 2, NIA2-1, NR, NITRATE REDUCTASE, NR2, NITRATE REDUCTASE 2 |
1.599 |
0.089 |
| 83 |
263691_at |
AT1G26880
|
[Ribosomal protein L34e superfamily protein] |
1.597 |
-0.029 |
| 84 |
256934_at |
AT3G22530
|
unknown |
1.596 |
0.139 |
| 85 |
249176_at |
AT5G42980
|
ATH3, thioredoxin H-type 3, ATTRX3, thioredoxin 3, ATTRXH3, TRX3, thioredoxin 3, TRXH3, THIOREDOXIN H3 |
1.593 |
0.026 |
| 86 |
251921_at |
AT3G53890
|
[Ribosomal protein S21e ] |
1.593 |
-0.098 |
| 87 |
263432_at |
AT2G22230
|
[Thioesterase superfamily protein] |
1.591 |
-0.136 |
| 88 |
263838_at |
AT2G36880
|
MAT3, methionine adenosyltransferase 3 |
1.589 |
-0.221 |
| 89 |
245155_at |
AT5G12470
|
unknown |
1.589 |
0.012 |
| 90 |
264344_at |
AT1G11910
|
APA1, aspartic proteinase A1, ATAPA1 |
1.588 |
0.035 |
| 91 |
263880_at |
AT2G21960
|
unknown |
1.588 |
-0.051 |
| 92 |
251664_at |
AT3G56940
|
ACSF, CHL27, CRD1, COPPER RESPONSE DEFECT 1 |
1.588 |
0.104 |
| 93 |
264668_at |
AT1G09780
|
iPGAM1, 2,3-biphosphoglycerate-independent phosphoglycerate mutase 1 |
1.588 |
-0.206 |
| 94 |
257938_at |
AT3G19820
|
CBB1, CABBAGE 1, DIM, DIMINUTIA, DIM1, DIMINUTO 1, DWF1, DWARF 1, EVE1, ENHANCED VERY-LOW-FLUENCE RESPONSES 1 |
1.588 |
-0.001 |
| 95 |
261379_at |
AT1G18720
|
unknown |
1.587 |
0.016 |
| 96 |
252325_at |
AT3G48560
|
AHAS, ACETOHYDROXY ACID SYNTHASE, ALS, ACETOLACTATE SYNTHASE, CSR1, chlorsulfuron/imidazolinone resistant 1, IMR1, IMIDAZOLE RESISTANT 1, TZP5, TRIAZOLOPYRIMIDINE RESISTANT 5 |
1.584 |
0.035 |
| 97 |
260308_at |
AT1G70610
|
ABCB26, ATP-binding cassette B26, ATTAP1, transporter associated with antigen processing protein 1, TAP1, transporter associated with antigen processing protein 1 |
1.580 |
0.358 |
| 98 |
258884_at |
AT3G10050
|
OMR1, L-O-methylthreonine resistant 1 |
1.579 |
-0.109 |
| 99 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.577 |
-0.391 |
| 100 |
255598_at |
AT4G00830
|
LIF2, LHP1-Interacting Factor 2 |
1.574 |
0.147 |
| 101 |
261410_at |
AT1G07610
|
MT1C, metallothionein 1C |
1.572 |
0.034 |
| 102 |
267526_at |
AT2G30570
|
PSBW, photosystem II reaction center W |
1.571 |
0.008 |
| 103 |
262163_at |
AT1G77940
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
1.569 |
-0.062 |
| 104 |
260651_at |
AT1G32460
|
unknown |
1.568 |
0.038 |
| 105 |
246745_at |
AT5G27770
|
[Ribosomal L22e protein family] |
1.568 |
-0.023 |
| 106 |
255447_at |
AT4G02790
|
EMB3129, EMBRYO DEFECTIVE 3129 |
1.567 |
-0.137 |
| 107 |
261122_at |
AT1G75330
|
OTC, ornithine carbamoyltransferase |
1.566 |
-0.019 |
| 108 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
1.563 |
0.594 |
| 109 |
267154_at |
AT2G30870
|
ATGSTF10, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 10, ATGSTF4, ERD13, EARLY DEHYDRATION-INDUCED 13, GSTF10, glutathione S-transferase PHI 10 |
1.562 |
0.030 |
| 110 |
259098_at |
AT3G04790
|
EMB3119, EMBRYO DEFECTIVE 3119 |
1.560 |
-0.171 |
| 111 |
253273_at |
AT4G34180
|
[Cyclase family protein] |
1.560 |
0.042 |
| 112 |
249826_at |
AT5G23310
|
FSD3, Fe superoxide dismutase 3 |
1.559 |
-0.317 |
| 113 |
260586_at |
AT2G43630
|
unknown |
1.556 |
0.382 |
| 114 |
257789_at |
AT3G27020
|
AtYSL6, YSL6, YELLOW STRIPE like 6 |
1.555 |
-0.055 |
| 115 |
247376_at |
AT5G63310
|
ATNDPK2, ARABIDOPSIS NUCLEOSIDE DIPHOSPHATE KINASE 2, NDPK IA, NUCLEOSIDE DIPHOSPHATE KINASE IA, NDPK IA IA, NDPK1A, NDP KINASE 1A, NDPK2, nucleoside diphosphate kinase 2 |
1.555 |
-0.165 |
| 116 |
261635_at |
AT1G50020
|
unknown |
1.555 |
0.098 |
| 117 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
1.554 |
-0.418 |
| 118 |
257753_at |
AT3G18740
|
RLK902, receptor-like kinase 902 |
1.553 |
-0.022 |
| 119 |
260258_at |
AT1G74270
|
[Ribosomal protein L35Ae family protein] |
1.553 |
-0.020 |
| 120 |
246035_at |
AT5G08300
|
[Succinyl-CoA ligase, alpha subunit] |
1.553 |
0.007 |
| 121 |
252561_at |
AT3G45980
|
H2B, HISTONE H2B, HTB9 |
1.551 |
-0.084 |
| 122 |
264903_at |
AT1G23190
|
PGM3, phosphoglucomutase 3 |
1.550 |
0.020 |
| 123 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
1.545 |
0.029 |
| 124 |
251157_at |
AT3G63140
|
CSP41A, chloroplast stem-loop binding protein of 41 kDa |
1.545 |
0.224 |
| 125 |
264837_at |
AT1G03600
|
PSB27 |
1.544 |
-0.064 |
| 126 |
254903_at |
AT4G11150
|
emb2448, embryo defective 2448, TUF, vacuolar ATP synthase subunit E1, TUFF, VHA-E1 |
1.544 |
0.040 |
| 127 |
253775_at |
AT4G28440
|
[Nucleic acid-binding, OB-fold-like protein] |
1.543 |
0.023 |
| 128 |
253920_at |
AT4G27230
|
HTA2, histone H2A 2 |
1.541 |
-0.277 |
| 129 |
256191_at |
AT1G30130
|
unknown |
1.541 |
0.097 |
| 130 |
249453_at |
AT5G39510
|
ATVTI11, VESICLE TRANSPORT V-SNARE 11, ATVTI1A, SGR4, SHOOT GRAVITROPSIM 4, VTI11, VESICLE TRANSPORT V-SNARE 11, VTI1A, ZIG, ZIG1 |
1.540 |
0.114 |
| 131 |
246454_at |
AT5G16710
|
DHAR3, dehydroascorbate reductase 1 |
1.538 |
-0.024 |
| 132 |
261355_at |
AT1G79750
|
ATNADP-ME4, Arabidopsis thaliana NADP-malic enzyme 4, NADP-ME4, NADP-malic enzyme 4 |
1.538 |
-0.025 |
| 133 |
262626_at |
AT1G06430
|
FTSH8, FTSH protease 8 |
1.536 |
0.231 |
| 134 |
254763_at |
AT4G13170
|
[Ribosomal protein L13 family protein] |
1.536 |
-0.104 |
| 135 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
1.536 |
0.051 |
| 136 |
261304_at |
AT1G48440
|
[B-cell receptor-associated 31-like] |
1.534 |
0.003 |
| 137 |
258811_at |
AT3G03990
|
[alpha/beta-Hydrolases superfamily protein] |
1.534 |
0.204 |
| 138 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
1.533 |
0.035 |
| 139 |
257983_at |
AT3G20790
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.533 |
-0.024 |
| 140 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
1.533 |
-0.014 |
| 141 |
246421_at |
AT5G16880
|
[Target of Myb protein 1] |
1.532 |
0.153 |
| 142 |
263256_at |
AT1G10500
|
ATCPISCA, chloroplast-localized ISCA-like protein, CPISCA, chloroplast-localized ISCA-like protein |
1.531 |
0.119 |
| 143 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
1.530 |
-0.174 |
| 144 |
255842_at |
AT2G33530
|
scpl46, serine carboxypeptidase-like 46 |
1.529 |
-0.204 |
| 145 |
245512_at |
AT4G15770
|
[RNA binding] |
1.528 |
-0.036 |
| 146 |
245263_at |
AT4G17740
|
[Peptidase S41 family protein] |
1.526 |
-0.217 |
| 147 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
1.526 |
0.457 |
| 148 |
256186_at |
AT1G51680
|
4CL.1, 4-COUMARATE:COA LIGASE 1, 4CL1, 4-coumarate:CoA ligase 1, AT4CL1, ARABIDOPSIS THALIANA 4-COUMARATE:COA LIGASE 1 |
1.525 |
-0.107 |
| 149 |
256785_at |
AT3G13720
|
PRA1.F3, PRENYLATED RAB ACCEPTOR 1.F3, PRA8 |
1.523 |
-0.140 |
| 150 |
263856_at |
AT2G04410
|
[RPM1-interacting protein 4 (RIN4) family protein] |
1.521 |
0.070 |
| 151 |
264007_at |
AT2G21140
|
ATPRP2, proline-rich protein 2, PRP2, proline-rich protein 2 |
1.521 |
-0.155 |
| 152 |
267294_at |
AT2G23670
|
YCF37, homolog of Synechocystis YCF37 |
1.517 |
-0.225 |
| 153 |
260940_at |
AT1G45000
|
[AAA-type ATPase family protein] |
1.516 |
0.026 |
| 154 |
252932_at |
AT4G39080
|
VHA-A3, vacuolar proton ATPase A3 |
1.516 |
-0.007 |
| 155 |
246730_at |
AT5G28060
|
[Ribosomal protein S24e family protein] |
1.511 |
0.051 |
| 156 |
258114_at |
AT3G14660
|
CYP72A13, cytochrome P450, family 72, subfamily A, polypeptide 13 |
1.511 |
0.031 |
| 157 |
252669_at |
AT3G44100
|
[MD-2-related lipid recognition domain-containing protein] |
1.511 |
0.004 |
| 158 |
261108_at |
AT1G62960
|
ACS10, ACC synthase 10 |
1.511 |
-0.132 |
| 159 |
263921_at |
AT2G36460
|
FBA6, fructose-bisphosphate aldolase 6 |
1.506 |
0.429 |
| 160 |
262908_at |
AT1G59900
|
AT-E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit, E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit |
1.506 |
0.065 |
| 161 |
246408_at |
AT1G57680
|
Cand1, candidate G-protein Coupled Receptor 1 |
1.505 |
0.142 |
| 162 |
258715_at |
AT3G09630
|
[Ribosomal protein L4/L1 family] |
1.504 |
-0.047 |
| 163 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
1.503 |
-0.143 |
| 164 |
245400_at |
AT4G17040
|
CLPR4, CLP protease R subunit 4, HON5, happy on norflurazon 5 |
1.502 |
0.004 |
| 165 |
248146_at |
AT5G54940
|
[Translation initiation factor SUI1 family protein] |
1.502 |
0.450 |
| 166 |
263755_at |
AT2G21340
|
[MATE efflux family protein] |
1.498 |
0.077 |
| 167 |
255483_at |
AT4G02500
|
ATXT2, ARABIDOPSIS THALIANA UDP-XYLOSYLTRANSFERASE 2, XT2, UDP-xylosyltransferase 2, XXT2, XYG XYLOSYLTRANSFERASE 2 |
1.497 |
-0.044 |
| 168 |
253705_at |
AT4G29130
|
ATHXK1, ARABIDOPSIS THALIANA HEXOKINASE 1, GIN2, GLUCOSE INSENSITIVE 2, HXK1, hexokinase 1 |
1.497 |
-0.133 |
| 169 |
262878_at |
AT1G64770
|
NDF2, NDH-dependent cyclic electron flow 1, NDH45, NAD(P)H DEHYDROGENASE SUBUNIT 45, PnsB2, Photosynthetic NDH subcomplex B 2 |
1.496 |
-0.089 |
| 170 |
257205_at |
AT3G16520
|
UGT88A1, UDP-glucosyl transferase 88A1 |
1.496 |
-0.078 |
| 171 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
1.494 |
0.466 |
| 172 |
247566_at |
AT5G61170
|
[Ribosomal protein S19e family protein] |
1.489 |
-0.157 |
| 173 |
253860_at |
AT4G27700
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
1.488 |
0.038 |
| 174 |
256226_at |
AT1G56280
|
ATDI19, drought-induced 19, DI19, drought-induced 19 |
1.487 |
0.125 |
| 175 |
266057_at |
AT2G40660
|
[Nucleic acid-binding, OB-fold-like protein] |
1.487 |
-0.013 |
| 176 |
246636_at |
AT5G34850
|
ATPAP26, PURPLE ACID PHOSPHATASE 26, PAP26, purple acid phosphatase 26 |
1.486 |
-0.007 |
| 177 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
1.483 |
0.052 |
| 178 |
249515_at |
AT5G38530
|
TSBtype2, tryptophan synthase beta type 2 |
1.481 |
-0.111 |
| 179 |
256228_at |
AT1G56190
|
[Phosphoglycerate kinase family protein] |
1.480 |
-0.048 |
| 180 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
1.479 |
-0.124 |
| 181 |
266552_at |
AT2G46330
|
AGP16, arabinogalactan protein 16, ATAGP16 |
1.479 |
-0.137 |
| 182 |
266521_at |
AT2G24020
|
[Uncharacterised BCR, YbaB family COG0718] |
1.477 |
0.056 |
| 183 |
249815_at |
AT5G23900
|
[Ribosomal protein L13e family protein] |
1.477 |
0.005 |
| 184 |
246146_at |
AT5G20050
|
[Protein kinase superfamily protein] |
1.477 |
0.027 |
| 185 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
1.474 |
0.174 |
| 186 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.474 |
-0.077 |
| 187 |
261789_at |
AT1G15930
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
1.472 |
-0.078 |
| 188 |
266495_at |
AT2G07050
|
CAS1, cycloartenol synthase 1 |
1.469 |
-0.077 |
| 189 |
258674_at |
AT3G08740
|
[elongation factor P (EF-P) family protein] |
1.469 |
-0.169 |
| 190 |
257880_at |
AT3G16910
|
AAE7, acyl-activating enzyme 7, ACN1, ACETATE NON-UTILIZING 1 |
1.469 |
0.084 |
| 191 |
253372_at |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
1.468 |
-0.381 |
| 192 |
253954_at |
AT4G26970
|
ACO2, aconitase 2 |
1.468 |
0.099 |
| 193 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
1.465 |
0.118 |
| 194 |
255633_at |
AT4G00585
|
unknown |
1.465 |
0.004 |
| 195 |
264286_at |
AT1G61870
|
PPR336, pentatricopeptide repeat 336 |
1.465 |
-0.185 |
| 196 |
247571_at |
AT5G61210
|
ATSNAP33, ATSNAP33B, SNAP33, soluble N-ethylmaleimide-sensitive factor adaptor protein 33, SNP33 |
1.465 |
0.098 |
| 197 |
248217_at |
AT5G53560
|
ATB5-A, ATCB5-E, ARABIDOPSIS CYTOCHROME B5 ISOFORM E, B5 #2, CB5-E, cytochrome B5 isoform E |
1.465 |
0.028 |
| 198 |
267251_at |
AT2G23070
|
[Protein kinase superfamily protein] |
1.464 |
0.109 |
| 199 |
262619_at |
AT1G06550
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
1.462 |
0.048 |
| 200 |
255411_at |
AT4G03110
|
AtBRN1, AtRBP-DR1, RNA-binding protein-defense related 1, BRN1, Bruno-like 1, RBP-DR1, RNA-binding protein-defense related 1 |
1.461 |
0.287 |
| 201 |
255199_at |
AT4G07390
|
[Mannose-P-dolichol utilization defect 1 protein] |
1.461 |
-0.048 |
| 202 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
1.460 |
-0.004 |
| 203 |
253386_at |
AT4G33030
|
SQD1, sulfoquinovosyldiacylglycerol 1 |
1.460 |
-0.018 |
| 204 |
251640_at |
AT3G57450
|
unknown |
1.459 |
0.132 |
| 205 |
258532_at |
AT3G06700
|
[Ribosomal L29e protein family] |
1.459 |
-0.059 |
| 206 |
257227_at |
AT3G27820
|
ATMDAR4, MONODEHYDROASCORBATE REDUCTASE 4, MDAR4, monodehydroascorbate reductase 4 |
1.456 |
0.012 |
| 207 |
253201_at |
AT4G34620
|
SSR16, small subunit ribosomal protein 16 |
1.456 |
0.021 |
| 208 |
257817_at |
AT3G25150
|
[Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain] |
1.455 |
0.084 |
| 209 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
1.455 |
-0.000 |
| 210 |
252410_at |
AT3G47450
|
ATNOA1, ATNOS1, NOA1, NO ASSOCIATED 1, NOS1, NITRIC OXIDE SYNTHASE 1, RIF1, RESISTANT TO INHIBITION WITH FOSMIDOMYCIN 1 |
1.455 |
-0.112 |
| 211 |
249231_at |
AT5G42030
|
ABIL4, ABL interactor-like protein 4 |
1.452 |
0.097 |
| 212 |
255982_at |
AT1G34000
|
OHP2, one-helix protein 2 |
1.451 |
0.037 |
| 213 |
260014_at |
AT1G68010
|
ATHPR1, HPR, hydroxypyruvate reductase |
1.450 |
0.087 |
| 214 |
266925_at |
AT2G45740
|
PEX11D, peroxin 11D |
1.450 |
-0.000 |
| 215 |
253858_at |
AT4G27600
|
NARA5, GENES NECESSARY FOR THE ACHIEVEMENT OF RUBISCO ACCUMULATION 5 |
1.448 |
-0.081 |
| 216 |
263987_at |
AT2G42690
|
[alpha/beta-Hydrolases superfamily protein] |
1.448 |
-0.101 |
| 217 |
247007_at |
AT5G67500
|
ATVDAC2, ARABIDOPSIS THALIANA VOLTAGE DEPENDENT ANION CHANNEL 2, VDAC2, voltage dependent anion channel 2 |
1.445 |
-0.024 |
| 218 |
263663_at |
AT1G04410
|
c-NAD-MDH1, cytosolic-NAD-dependent malate dehydrogenase 1 |
1.445 |
-0.071 |
| 219 |
262985_s_at |
AT1G23290
|
RPL27A, RIBOSOMAL PROTEIN L27A, RPL27AB |
1.444 |
-0.092 |
| 220 |
267470_at |
AT2G30490
|
ATC4H, CINNAMATE 4-HYDROXYLASE, C4H, cinnamate-4-hydroxylase, CYP73A5, REF3, REDUCED EPRDERMAL FLUORESCENCE 3 |
1.444 |
0.008 |
| 221 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
1.443 |
-0.199 |
| 222 |
257731_at |
AT3G18430
|
[Calcium-binding EF-hand family protein] |
1.441 |
0.051 |
| 223 |
260746_at |
AT1G78380
|
ATGSTU19, A. THALIANA GLUTATHIONE S-TRANSFERASE TAU 19, GST8, GLUTATHIONE TRANSFERASE 8, GSTU19, glutathione S-transferase TAU 19 |
1.440 |
0.210 |
| 224 |
260487_at |
AT1G51510
|
Y14 |
1.439 |
0.034 |
| 225 |
252318_at |
AT3G48730
|
GSA2, glutamate-1-semialdehyde 2,1-aminomutase 2 |
1.439 |
-0.236 |
| 226 |
260501_at |
AT2G41770
|
unknown |
1.437 |
-0.159 |
| 227 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
1.437 |
0.131 |
| 228 |
246744_at |
AT5G27760
|
[Hypoxia-responsive family protein] |
1.435 |
0.123 |
| 229 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
1.435 |
0.136 |
| 230 |
254616_at |
AT4G18710
|
ATSK21, SHAGGY-LIKE KINASE 21, BIN2, BRASSINOSTEROID-INSENSITIVE 2, DWF12, DWARF 12, SK21, SHAGGY-LIKE KINASE 21, UCU1, ULTRACURVATA 1 |
1.432 |
0.008 |
| 231 |
246301_at |
AT3G51850
|
CPK13, calcium-dependent protein kinase 13 |
1.432 |
-0.019 |
| 232 |
249810_at |
AT5G23920
|
unknown |
1.430 |
-0.032 |
| 233 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
1.430 |
0.012 |
| 234 |
262411_at |
AT1G34640
|
[peptidases] |
1.429 |
-0.135 |
| 235 |
252849_at |
AT4G40040
|
H3.3, histone 3.3 |
1.429 |
0.006 |
| 236 |
252027_at |
AT3G52850
|
ATELP, ARABIDOPSIS THALIANA EPIDERMAL GROWTH FACTOR RECEPTOR-LIKE PROTEIN, ATELP1, ATVSR1, BP-80, BP80, BP80-1;1, binding protein of 80 kDa 1;1, BP80B, GFS1, Green fluorescent seed 1, VSR1, vacuolar sorting receptor homolog 1, VSR1;1, VACUOLAR SORTING RECEPTOR 1;1 |
1.429 |
0.006 |
| 237 |
262104_at |
AT1G02910
|
LPA1, LOW PSII ACCUMULATION1 |
1.429 |
0.202 |
| 238 |
252935_at |
AT4G39150
|
[DNAJ heat shock N-terminal domain-containing protein] |
1.428 |
0.027 |
| 239 |
251413_at |
AT3G60320
|
unknown |
1.428 |
0.171 |
| 240 |
258226_at |
AT3G15730
|
PLD, PLDALPHA1, phospholipase D alpha 1 |
1.426 |
-0.000 |
| 241 |
266123_at |
AT2G45180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.426 |
-0.013 |
| 242 |
254669_at |
AT4G18370
|
DEG5, degradation of periplasmic proteins 5, DEGP5, DEGP PROTEASE 5, HHOA, PROTEASE HHOA PRECUSOR |
1.426 |
-0.099 |
| 243 |
251801_at |
AT3G55440
|
ATCTIMC, CYTOSOLIC TRIOSE PHOSPHATE ISOMERASE, CYTOTPI, CYTOSOLIC ISOFORM TRIOSE PHOSPHATE ISOMERASE, TPI, triosephosphate isomerase |
1.425 |
-0.020 |
| 244 |
259521_at |
AT1G12410
|
CLP2, CLP protease proteolytic subunit 2, CLPR2, EMB3146, EMBRYO DEFECTIVE 3146, NCLPP2, NUCLEAR-ENCODED CLP PROTEASE P2 |
1.425 |
0.010 |
| 245 |
250922_at |
AT5G03345
|
unknown |
1.425 |
0.061 |
| 246 |
266289_at |
AT2G29390
|
ATSMO2, STEROL 4-ALPHA-METHYL-OXIDASE 2, ATSMO2-2, Arabidopsis thaliana sterol 4-alpha-methyl-oxidase 2-2, SMO2-2, sterol 4-alpha-methyl-oxidase 2-2 |
1.424 |
-0.043 |
| 247 |
257228_at |
AT3G27890
|
NQR, NADPH:quinone oxidoreductase |
1.423 |
0.020 |
| 248 |
252998_at |
AT4G38510
|
AtVAB2, VAB2, V-ATPase B subunit 2 |
1.423 |
-0.039 |
| 249 |
247101_at |
AT5G66530
|
[Galactose mutarotase-like superfamily protein] |
1.423 |
-0.114 |
| 250 |
245832_at |
AT1G48850
|
EMB1144, embryo defective 1144 |
1.423 |
-0.026 |
| 251 |
264970_at |
AT1G67280
|
[Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily protein] |
1.422 |
0.052 |
| 252 |
254623_at |
AT4G18480
|
CH-42, CHLORINA 42, CH42, CHLORINA 42, CHL11, CHLI-1, CHLI1 |
1.422 |
-0.240 |
| 253 |
256034_at |
AT1G07080
|
[Thioredoxin superfamily protein] |
1.422 |
-0.034 |
| 254 |
260409_at |
AT1G69935
|
SHW1, SHORT HYPOCOTYL IN WHITE LIGHT1 |
1.421 |
0.100 |
| 255 |
255263_at |
AT4G05160
|
[AMP-dependent synthetase and ligase family protein] |
1.421 |
-0.006 |
| 256 |
256207_at |
AT1G50920
|
[Nucleolar GTP-binding protein] |
1.420 |
0.014 |
| 257 |
265738_at |
AT2G01350
|
QPT, quinolinate phoshoribosyltransferase |
1.420 |
0.069 |
| 258 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
1.419 |
0.082 |
| 259 |
264422_at |
AT1G43130
|
LCV2, like COV 2 |
1.419 |
0.110 |
| 260 |
248962_at |
AT5G45680
|
ATFKBP13, FK506 BINDING PROTEIN 13, FKBP13, FK506-binding protein 13 |
1.418 |
-0.267 |
| 261 |
256145_at |
AT1G48750
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.418 |
-0.108 |
| 262 |
259161_at |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
1.417 |
0.112 |
| 263 |
251905_at |
AT3G53710
|
AGD6, ARF-GAP domain 6 |
1.415 |
0.259 |
| 264 |
262426_s_at |
AT1G47640
|
unknown |
1.415 |
0.121 |
| 265 |
254117_at |
AT4G24750
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
1.415 |
-0.344 |
| 266 |
256383_at |
AT1G66820
|
[glycine-rich protein] |
1.415 |
-0.287 |
| 267 |
245525_at |
AT4G15930
|
[Dynein light chain type 1 family protein] |
1.415 |
0.299 |
| 268 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
1.415 |
-0.244 |
| 269 |
255623_at |
AT4G01310
|
[Ribosomal L5P family protein] |
1.415 |
-0.060 |
| 270 |
257911_at |
AT3G25530
|
ATGHBDH, AtGLYR1, GHBDH, GLYR1, glyoxylate reductase 1, GR1, GLYOXYLATE REDUCTASE 1 |
1.414 |
-0.006 |
| 271 |
256411_at |
AT1G66670
|
CLPP3, CLP protease proteolytic subunit 3, NCLPP3 |
1.414 |
-0.063 |
| 272 |
248126_at |
AT5G54760
|
[Translation initiation factor SUI1 family protein] |
1.413 |
0.090 |
| 273 |
253811_at |
AT4G28190
|
ULT, ULTRAPETALA, ULT1, ULTRAPETALA1 |
1.412 |
0.057 |
| 274 |
262824_at |
AT1G11650
|
ATRBP45B, RBP45B |
1.412 |
0.083 |
| 275 |
264307_at |
AT1G61900
|
unknown |
1.412 |
0.125 |
| 276 |
263728_at |
AT1G60070
|
[Adaptor protein complex AP-1, gamma subunit] |
1.412 |
-0.021 |
| 277 |
249007_at |
AT5G44650
|
AtCEST, Arabidopsis thaliana chloroplast protein-enhancing stress tolerance, CEST, chloroplast protein-enhancing stress tolerance, Y3IP1, Ycf3-interacting protein 1 |
1.412 |
-0.275 |
| 278 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
1.411 |
-0.092 |
| 279 |
255451_at |
AT4G02860
|
[Phenazine biosynthesis PhzC/PhzF protein] |
1.410 |
-0.015 |
| 280 |
260507_at |
AT1G47200
|
WPP2, WPP domain protein 2 |
1.409 |
-0.053 |
| 281 |
251667_at |
AT3G57150
|
AtCBF5, AtNAP57, CBF5, NAP57, homologue of NAP57 |
1.408 |
-0.089 |
| 282 |
248838_at |
AT5G46800
|
BOU, A BOUT DE SOUFFLE |
1.407 |
0.034 |
| 283 |
258790_at |
AT3G04610
|
FLK, flowering locus KH domain |
1.406 |
0.061 |
| 284 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
1.406 |
0.036 |
| 285 |
259664_at |
AT1G55330
|
AGP21, arabinogalactan protein 21, ATAGP21, ARABINOGALACTAN PROTEIN 21 |
1.405 |
0.121 |
| 286 |
255674_at |
AT4G00430
|
PIP1;4, plasma membrane intrinsic protein 1;4, PIP1E, PLASMA MEMBRANE INTRINSIC PROTEIN 1E, TMP-C, TRANSMEMBRANE PROTEIN C |
1.404 |
0.259 |
| 287 |
259847_at |
AT1G72170
|
unknown |
1.403 |
-0.041 |
| 288 |
266509_at |
AT2G47940
|
DEG2, degradation of periplasmic proteins 2, DEGP2, DEGP protease 2, EMB3117, EMBRYO DEFECTIVE 3117 |
1.403 |
-0.183 |
| 289 |
266608_at |
AT2G35500
|
SKL2, shikimate kinase-like 2 |
1.403 |
-0.135 |
| 290 |
266285_at |
AT2G29180
|
unknown |
1.403 |
-0.174 |
| 291 |
257023_at |
AT3G19760
|
EIF4A-III, eukaryotic initiation factor 4A-III |
1.402 |
0.140 |
| 292 |
258970_at |
AT3G10410
|
CPY, CARBOXYPEPTIDASE Y, SCPL49, SERINE CARBOXYPEPTIDASE-LIKE 49 |
1.402 |
-0.084 |
| 293 |
262161_at |
AT1G52600
|
[Peptidase S24/S26A/S26B/S26C family protein] |
1.401 |
0.109 |
| 294 |
263669_at |
AT1G04400
|
AT-PHH1, ATCRY2, CRY2, cryptochrome 2, FHA, PHH1 |
1.400 |
0.121 |
| 295 |
264724_at |
AT1G22920
|
AJH1, ARABIDOPSIS JAB1 HOMOLOG 1, CSN5A, COP9 signalosome 5A |
1.400 |
-0.024 |
| 296 |
260511_at |
AT1G51570
|
Calcium-dependent lipid-binding (CaLB domain) plant phosphoribosyltransferase family protein |
1.399 |
-0.127 |
| 297 |
252033_at |
AT3G51950
|
[Zinc finger (CCCH-type) family protein / RNA recognition motif (RRM)-containing protein] |
1.399 |
-0.084 |
| 298 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
1.399 |
0.084 |
| 299 |
256879_at |
AT3G26370
|
[O-fucosyltransferase family protein] |
1.399 |
0.025 |
| 300 |
250863_at |
AT5G04750
|
[F1F0-ATPase inhibitor protein, putative] |
1.398 |
0.232 |
| 301 |
263781_at |
AT2G46360
|
unknown |
-1.866 |
0.082 |
| 302 |
251159_at |
AT3G63230
|
unknown |
-1.849 |
0.198 |
| 303 |
251114_at |
AT5G01380
|
[Homeodomain-like superfamily protein] |
-1.824 |
0.554 |
| 304 |
258734_at |
AT3G05860
|
[MADS-box transcription factor family protein] |
-1.822 |
0.156 |
| 305 |
252216_at |
AT3G50420
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-1.809 |
0.130 |
| 306 |
252696_at |
AT3G43650
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.0e-43 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-1.808 |
0.154 |
| 307 |
258028_at |
AT3G27473
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.789 |
0.120 |
| 308 |
254895_at |
AT4G11870
|
unknown |
-1.785 |
0.208 |
| 309 |
267109_at |
AT2G14810
|
unknown |
-1.785 |
0.102 |
| 310 |
262350_at |
AT2G48150
|
ATGPX4, glutathione peroxidase 4, GPX4, glutathione peroxidase 4 |
-1.782 |
0.211 |
| 311 |
263682_at |
AT1G26860
|
unknown |
-1.764 |
0.271 |
| 312 |
267575_at |
AT2G30690
|
unknown |
-1.754 |
0.261 |
| 313 |
248431_at |
AT5G51470
|
[Auxin-responsive GH3 family protein] |
-1.751 |
0.101 |
| 314 |
260115_at |
AT1G33870
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.748 |
0.114 |
| 315 |
250594_at |
AT5G07760
|
[formin homology 2 domain-containing protein / FH2 domain-containing protein] |
-1.733 |
0.018 |
| 316 |
252460_at |
AT3G47230
|
unknown |
-1.733 |
0.171 |
| 317 |
248314_at |
AT5G52620
|
[F-box associated ubiquitination effector family protein] |
-1.728 |
0.127 |
| 318 |
252739_at |
AT3G43250
|
unknown |
-1.725 |
0.172 |
| 319 |
253936_at |
AT4G26880
|
[Stigma-specific Stig1 family protein] |
-1.725 |
0.056 |
| 320 |
255343_at |
AT4G04530
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT1G27780.1)] |
-1.724 |
0.072 |
| 321 |
264139_at |
AT1G78940
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-1.717 |
0.050 |
| 322 |
256487_at |
AT1G31540
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-1.706 |
0.094 |
| 323 |
259086_at |
AT3G04990
|
unknown |
-1.705 |
0.104 |
| 324 |
265167_at |
AT1G23680
|
unknown |
-1.701 |
0.037 |
| 325 |
265982_at |
AT2G11190
|
[copia-like retrotransposon family, has a 3.5e-90 P-value blast match to GB:CAA32025 ORF (Ty1_Copia-element) (Nicotiana tabacum)GB:CAA32025 ORF (Ty1_Copia-element) (Nicotiana tabacum)] |
-1.701 |
0.133 |
| 326 |
246661_at |
AT5G35280
|
unknown |
-1.697 |
0.169 |
| 327 |
256635_at |
AT3G28260
|
unknown |
-1.687 |
0.102 |
| 328 |
247369_at |
AT5G63340
|
unknown |
-1.686 |
0.125 |
| 329 |
254856_at |
AT4G12160
|
[pseudogene] |
-1.679 |
0.123 |
| 330 |
266393_at |
AT2G41260
|
ATM17, M17 |
-1.677 |
0.157 |
| 331 |
253902_at |
AT4G27170
|
AT2S4, SESA4, seed storage albumin 4 |
-1.671 |
0.145 |
| 332 |
254589_at |
AT4G18850
|
unknown |
-1.659 |
0.088 |
| 333 |
266773_at |
AT2G29040
|
[Exostosin family protein] |
-1.655 |
0.178 |
| 334 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
-1.646 |
0.492 |
| 335 |
257961_at |
AT3G19780
|
[LOCATED IN: endomembrane system] |
-1.643 |
0.125 |
| 336 |
255242_at |
AT4G05610
|
[pseudogene] |
-1.642 |
0.102 |
| 337 |
259108_at |
AT3G05540
|
[Methionine sulfoxide reductase (MSS4-like) family protein] |
-1.638 |
0.141 |
| 338 |
255138_at |
AT4G08380
|
[Proline-rich extensin-like family protein] |
-1.636 |
0.148 |
| 339 |
266966_at |
AT2G39520
|
unknown |
-1.629 |
0.149 |
| 340 |
247732_at |
AT5G59600
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-1.626 |
-0.013 |
| 341 |
256540_at |
AT3G32917
|
[gypsy-like retrotransposon family, has a 1.0e-114 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
-1.626 |
0.173 |
| 342 |
255653_at |
AT4G00960
|
[Protein kinase superfamily protein] |
-1.617 |
0.121 |
| 343 |
257316_at |
AT3G30750
|
unknown |
-1.617 |
0.227 |
| 344 |
248999_at |
AT5G44970
|
[Protein with RNI-like/FBD-like domains] |
-1.615 |
0.083 |
| 345 |
263745_at |
AT2G21450
|
CHR34, chromatin remodeling 34 |
-1.613 |
0.188 |
| 346 |
248859_at |
AT5G46660
|
[protein kinase C-like zinc finger protein] |
-1.612 |
0.014 |
| 347 |
256987_at |
AT3G28560
|
[BCS1 AAA-type ATPase] |
-1.610 |
0.109 |
| 348 |
245829_at |
AT1G57780
|
[heavy-metal-associated domain-containing protein] |
-1.607 |
0.179 |
| 349 |
251003_at |
AT5G02690
|
unknown |
-1.607 |
0.040 |
| 350 |
245177_at |
AT5G12380
|
ANNAT8, annexin 8 |
-1.605 |
0.136 |
| 351 |
245402_at |
AT4G17585
|
[Aluminium activated malate transporter family protein] |
-1.601 |
0.183 |
| 352 |
247905_at |
AT5G57400
|
unknown |
-1.597 |
0.246 |
| 353 |
261045_at |
AT1G01310
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-1.597 |
0.127 |
| 354 |
248383_at |
AT5G51900
|
[Cytochrome P450 family protein] |
-1.597 |
0.136 |
| 355 |
266645_at |
AT2G29880
|
unknown |
-1.595 |
0.147 |
| 356 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
-1.594 |
1.871 |
| 357 |
265838_at |
AT2G14550
|
[pseudogene] |
-1.594 |
0.229 |
| 358 |
248297_at |
AT5G53100
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.584 |
0.144 |
| 359 |
256180_at |
AT1G51810
|
[Leucine-rich repeat protein kinase family protein] |
-1.584 |
0.086 |
| 360 |
246925_at |
AT5G25180
|
CYP71B14, cytochrome P450, family 71, subfamily B, polypeptide 14 |
-1.583 |
0.196 |
| 361 |
251132_at |
AT5G01200
|
[Duplicated homeodomain-like superfamily protein] |
-1.583 |
0.235 |
| 362 |
253070_at |
AT4G37850
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-1.581 |
0.046 |
| 363 |
266144_at |
AT2G12170
|
unknown |
-1.580 |
0.128 |
| 364 |
261645_at |
AT1G27790
|
unknown |
-1.580 |
0.461 |
| 365 |
265515_at |
AT2G06120
|
unknown |
-1.577 |
0.134 |
| 366 |
265406_at |
AT2G16610
|
[CACTA-like transposase family (En/Spm), has a 6.1e-89 P-value blast match to GB:BAA20532 ORF of transposon Tdc1 (CACTA-element) (Daucus carota)] |
-1.577 |
0.062 |
| 367 |
251329_at |
AT3G61450
|
ATSYP73, SYP73, syntaxin of plants 73 |
-1.576 |
0.057 |
| 368 |
259886_at |
AT1G76370
|
[Protein kinase superfamily protein] |
-1.575 |
0.139 |
| 369 |
260934_at |
AT1G45100
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-1.575 |
0.143 |
| 370 |
264281_at |
AT1G61830
|
[pseudogene] |
-1.574 |
0.151 |
| 371 |
253937_at |
AT4G26890
|
MAPKKK16, mitogen-activated protein kinase kinase kinase 16 |
-1.573 |
0.121 |
| 372 |
265176_at |
AT1G23520
|
unknown |
-1.573 |
0.147 |
| 373 |
264500_at |
AT1G09370
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.568 |
0.138 |
| 374 |
251180_at |
AT3G62640
|
unknown |
-1.568 |
0.095 |
| 375 |
251715_at |
AT3G56390
|
unknown |
-1.567 |
0.116 |
| 376 |
263832_at |
AT2G40310
|
[Pectin lyase-like superfamily protein] |
-1.564 |
0.248 |
| 377 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
-1.564 |
0.456 |
| 378 |
263272_at |
AT2G11650
|
[pseudogene] |
-1.564 |
0.087 |
| 379 |
266754_at |
AT2G46980
|
ASY3, ASYNAPTIC 3, AtASY3, Arabidopsis thaliana ASYNAPTIC 3 |
-1.563 |
0.215 |
| 380 |
260994_at |
AT1G12130
|
[Flavin-binding monooxygenase family protein] |
-1.561 |
0.031 |
| 381 |
246823_at |
AT5G26970
|
unknown |
-1.560 |
0.157 |
| 382 |
265904_at |
AT2G25630
|
BGLU14, beta glucosidase 14 |
-1.558 |
0.120 |
| 383 |
252734_at |
AT3G43160
|
MEE38, maternal effect embryo arrest 38 |
-1.556 |
0.265 |
| 384 |
253924_at |
AT4G27110
|
COBL11, COBRA-like protein 11 precursor |
-1.553 |
0.091 |
| 385 |
250956_at |
AT5G03210
|
AtDIP2, DIP2, DBP-interacting protein 2 |
-1.547 |
0.353 |
| 386 |
252581_at |
AT3G45500
|
unknown |
-1.543 |
0.070 |
| 387 |
245836_at |
AT1G42250
|
unknown |
-1.543 |
0.138 |
| 388 |
245782_at |
AT1G35200
|
[pseudogene] |
-1.542 |
0.102 |
| 389 |
260713_at |
AT1G17615
|
[Disease resistance protein (TIR-NBS class)] |
-1.536 |
0.134 |
| 390 |
256584_at |
AT3G28750
|
unknown |
-1.534 |
0.122 |
| 391 |
253322_at |
AT4G33980
|
unknown |
-1.534 |
0.773 |
| 392 |
261565_at |
AT1G33220
|
[Glycosyl hydrolase superfamily protein] |
-1.533 |
0.067 |
| 393 |
257491_at |
AT1G06170
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-1.533 |
0.024 |
| 394 |
256695_at |
AT3G32090
|
[WRKY family transcription factor] |
-1.532 |
0.132 |
| 395 |
248930_at |
AT5G46010
|
[Homeodomain-like superfamily protein] |
-1.532 |
0.091 |
| 396 |
257682_at |
AT3G13240
|
unknown |
-1.531 |
0.171 |
| 397 |
249352_at |
AT5G40430
|
AtMYB22, myb domain protein 22, MYB22, myb domain protein 22 |
-1.527 |
0.146 |
| 398 |
258639_at |
AT3G07820
|
[Pectin lyase-like superfamily protein] |
-1.524 |
0.166 |
| 399 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
-1.523 |
0.144 |
| 400 |
263234_at |
AT1G10417
|
unknown |
-1.523 |
0.125 |
| 401 |
249643_at |
AT5G36990
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 8.5e-61 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
-1.522 |
0.104 |
| 402 |
262895_at |
AT1G59800
|
[Cullin family protein] |
-1.521 |
0.108 |
| 403 |
245922_at |
AT5G28810
|
unknown |
-1.516 |
0.123 |
| 404 |
248182_at |
AT5G54030
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.514 |
0.045 |
| 405 |
253503_at |
AT4G31950
|
CYP82C3, cytochrome P450, family 82, subfamily C, polypeptide 3 |
-1.514 |
0.038 |
| 406 |
251452_at |
AT3G60060
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.512 |
0.284 |
| 407 |
263407_at |
AT2G04038
|
AtbZIP48, basic leucine-zipper 48, bZIP48, basic leucine-zipper 48 |
-1.511 |
0.267 |
| 408 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
-1.511 |
0.061 |
| 409 |
253724_at |
AT4G29285
|
LCR24, low-molecular-weight cysteine-rich 24 |
-1.510 |
0.043 |
| 410 |
246688_at |
AT5G34450
|
unknown |
-1.509 |
0.063 |
| 411 |
257133_at |
AT3G17190
|
unknown |
-1.509 |
0.163 |
| 412 |
253504_at |
AT4G31960
|
unknown |
-1.509 |
0.086 |
| 413 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-1.509 |
-0.008 |
| 414 |
261797_at |
AT1G30455
|
[transcription regulators] |
-1.508 |
0.154 |
| 415 |
265541_at |
AT2G15800
|
transposable element gene |
-1.508 |
0.150 |
| 416 |
260828_at |
AT1G06750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.506 |
0.208 |
| 417 |
265303_at |
AT2G14040
|
[gypsy-like retrotransposon family (Athila), has a 1.0e-99 P-value blast match to gb|AAL06421.1|AF378079_1 reverse transcriptase (Athila4) (Arabidopsis thaliana) (Gypsy_Ty3-family)] |
-1.505 |
0.311 |
| 418 |
262294_at |
AT1G27610
|
unknown |
-1.504 |
0.182 |
| 419 |
252838_at |
AT3G42090
|
[contains domain LIN-9 RELATED (PTHR21689)] |
-1.503 |
0.101 |
| 420 |
263423_at |
AT2G31700
|
unknown |
-1.502 |
0.138 |
| 421 |
245923_at |
AT5G28820
|
unknown |
-1.502 |
0.106 |
| 422 |
259483_at |
AT1G43980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-1.500 |
0.089 |
| 423 |
263518_at |
AT2G21655
|
unknown |
-1.500 |
0.129 |
| 424 |
263615_at |
AT2G25230
|
AtMYB100, myb domain protein 100, MYB100, myb domain protein 100 |
-1.500 |
0.075 |
| 425 |
252577_at |
AT3G45460
|
[IBR domain containing protein] |
-1.499 |
0.145 |
| 426 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
-1.499 |
0.867 |
| 427 |
259223_at |
AT3G03660
|
WOX11, WUSCHEL related homeobox 11 |
-1.497 |
0.159 |
| 428 |
260212_at |
AT1G74480
|
AtRKD2, RKD2, RWP-RK domain containing 2, URP4, UAS-TAGGED ROOT PATTERNING4 |
-1.496 |
0.203 |
| 429 |
256295_at |
AT1G69470
|
unknown |
-1.493 |
0.107 |
| 430 |
257391_at |
AT2G32050
|
unknown |
-1.492 |
0.106 |
| 431 |
264888_at |
AT1G23070
|
unknown |
-1.492 |
0.144 |
| 432 |
265631_at |
AT2G14300
|
[pseudogene] |
-1.491 |
0.064 |
| 433 |
251382_at |
AT3G60730
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-1.491 |
0.098 |
| 434 |
264981_at |
AT1G27160
|
[valyl-tRNA synthetase / valine--tRNA ligase-related] |
-1.489 |
0.154 |
| 435 |
248479_at |
AT5G50910
|
unknown |
-1.489 |
0.144 |
| 436 |
255156_at |
AT4G07780
|
[gypsy-like retrotransposon family (Athila), has a 3.1e-225 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-1.488 |
0.050 |
| 437 |
256843_at |
AT3G31940
|
unknown |
-1.488 |
0.058 |
| 438 |
265972_at |
AT2G11235
|
[copia-like retrotransposon family, has a 1.6e-94 P-value blast match to gb|AAG52949.1| gag/pol polyprotein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
-1.488 |
0.118 |
| 439 |
250871_at |
AT5G03930
|
unknown |
-1.487 |
0.176 |
| 440 |
255282_at |
AT4G04600
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
-1.486 |
0.169 |
| 441 |
263888_at |
AT2G37000
|
[TCP family transcription factor ] |
-1.486 |
0.121 |
| 442 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
-1.486 |
0.087 |
| 443 |
264104_at |
AT2G13750
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.5e-17 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-1.485 |
0.118 |
| 444 |
257681_at |
AT3G13370
|
unknown |
-1.485 |
0.130 |
| 445 |
257186_at |
AT3G13130
|
unknown |
-1.484 |
0.113 |
| 446 |
266245_at |
AT2G27700
|
[eukaryotic translation initiation factor 2 family protein / eIF-2 family protein] |
-1.484 |
0.078 |
| 447 |
259046_at |
AT3G03400
|
[EF hand calcium-binding protein family] |
-1.482 |
0.169 |
| 448 |
258103_at |
AT3G23630
|
ATIPT7, ARABIDOPSIS THALIANA ISOPENTENYLTRANSFERASE 7, IPT7, isopentenyltransferase 7 |
-1.482 |
0.014 |
| 449 |
252219_at |
AT3G50160
|
unknown |
-1.478 |
0.086 |
| 450 |
261836_at |
AT1G16090
|
WAKL7, wall associated kinase-like 7 |
-1.474 |
0.209 |
| 451 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-1.473 |
0.068 |
| 452 |
264110_at |
AT2G13730
|
transposable element gene |
-1.473 |
0.155 |
| 453 |
265055_at |
AT1G52020
|
[pseudogene] |
-1.472 |
0.130 |
| 454 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
-1.470 |
0.357 |
| 455 |
254696_at |
AT4G17930
|
unknown |
-1.468 |
0.167 |
| 456 |
249883_at |
AT5G22900
|
ATCHX3, ARABIDOPSIS THALIANA CATION/H+ EXCHANGER 3, CHX3, cation/H+ exchanger 3 |
-1.467 |
0.108 |
| 457 |
247526_at |
AT5G61470
|
[C2H2-like zinc finger protein] |
-1.465 |
0.198 |
| 458 |
267554_at |
AT2G32790
|
[Ubiquitin-conjugating enzyme family protein] |
-1.464 |
0.119 |
| 459 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
-1.463 |
0.154 |
| 460 |
247581_at |
AT5G61350
|
[Protein kinase superfamily protein] |
-1.460 |
-0.121 |
| 461 |
259870_at |
AT1G76780
|
[HSP20-like chaperones superfamily protein] |
-1.458 |
0.890 |
| 462 |
260977_at |
AT1G53420
|
[Leucine-rich repeat transmembrane protein kinase] |
-1.457 |
0.123 |
| 463 |
255357_at |
AT4G03930
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-1.457 |
0.168 |
| 464 |
255122_at |
AT4G08490
|
[gypsy-like retrotransposon family (Athila), has a 4.7e-81 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-1.456 |
0.087 |
| 465 |
262662_at |
AT1G13920
|
[Remorin family protein] |
-1.455 |
0.014 |
| 466 |
263080_at |
AT2G12610
|
unknown |
-1.455 |
0.065 |
| 467 |
248215_at |
AT5G53680
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-1.454 |
1.799 |
| 468 |
247469_at |
AT5G62165
|
AGL42, AGAMOUS-like 42, FYF, FOREVER YOUNG FLOWER |
-1.453 |
0.055 |
| 469 |
263748_at |
AT2G21480
|
[Malectin/receptor-like protein kinase family protein] |
-1.450 |
0.186 |
| 470 |
249839_at |
AT5G23405
|
[HMG-box (high mobility group) DNA-binding family protein] |
-1.450 |
0.036 |
| 471 |
264706_at |
AT1G09720
|
NET2B, Networked 2B |
-1.448 |
0.092 |
| 472 |
251913_at |
AT3G53910
|
[malate dehydrogenase-related] |
-1.446 |
0.132 |
| 473 |
245068_at |
AT2G23260
|
UGT84B1, UDP-glucosyl transferase 84B1 |
-1.444 |
0.120 |
| 474 |
257468_at |
AT1G47470
|
unknown |
-1.443 |
0.148 |
| 475 |
248355_at |
AT5G52340
|
ATEXO70A2, exocyst subunit exo70 family protein A2, EXO70A2, exocyst subunit exo70 family protein A2 |
-1.443 |
0.170 |
| 476 |
263940_at |
AT2G35890
|
CPK25, calcium-dependent protein kinase 25 |
-1.442 |
0.091 |
| 477 |
253258_at |
AT4G34400
|
[AP2/B3-like transcriptional factor family protein] |
-1.442 |
0.126 |
| 478 |
249349_at |
AT5G40350
|
AtMYB24, myb domain protein 24, MYB24, myb domain protein 24 |
-1.439 |
0.204 |
| 479 |
259693_at |
AT1G63060
|
unknown |
-1.438 |
0.072 |
| 480 |
245664_at |
AT1G28327
|
unknown |
-1.438 |
0.136 |
| 481 |
264791_at |
AT2G17845
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.438 |
0.135 |
| 482 |
245157_at |
AT2G33160
|
NMA, NIMNA (Sanskrit for 'sunken or low) |
-1.437 |
0.123 |
| 483 |
251135_at |
AT5G01280
|
unknown |
-1.437 |
0.133 |
| 484 |
248194_at |
AT5G54095
|
unknown |
-1.436 |
0.056 |
| 485 |
255195_at |
AT4G07425
|
[CACTA-like transposase family (Tnp1/En/Spm), has a 2.5e-22 P-value blast match to ref|NP_189784.1| TNP1-related protein (Arabidopsis thaliana) (CACTA-element)] |
-1.435 |
0.151 |
| 486 |
262763_at |
AT1G28690
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-1.435 |
0.071 |
| 487 |
248876_at |
AT5G46120
|
unknown |
-1.434 |
0.136 |
| 488 |
248892_at |
AT5G46300
|
unknown |
-1.434 |
0.124 |
| 489 |
265198_at |
AT2G36760
|
UGT73C2, UDP-glucosyl transferase 73C2 |
-1.433 |
0.108 |
| 490 |
247090_at |
AT5G66370
|
unknown |
-1.433 |
0.056 |
| 491 |
248894_at |
AT5G46320
|
[MADS-box family protein] |
-1.432 |
0.068 |
| 492 |
252140_at |
AT3G51070
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.432 |
0.163 |
| 493 |
261997_at |
AT1G33817
|
[copia-like retrotransposon family, has a 8.1e-100 P-value blast match to gb|AAO73521.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-1.431 |
0.062 |
| 494 |
256559_at |
AT3G31340
|
[gypsy-like retrotransposon family (Athila), has a 7.3e-111 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-1.431 |
0.130 |
| 495 |
251755_at |
AT3G55790
|
unknown |
-1.429 |
0.031 |
| 496 |
249819_at |
AT5G23640
|
unknown |
-1.428 |
0.092 |
| 497 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
-1.426 |
-0.176 |
| 498 |
260846_at |
AT1G17300
|
unknown |
-1.425 |
0.195 |
| 499 |
248946_at |
AT5G45530
|
unknown |
-1.423 |
0.121 |
| 500 |
258144_at |
AT3G18180
|
[Glycosyltransferase family 61 protein] |
-1.420 |
0.075 |
| 501 |
248317_at |
AT5G52680
|
[Copper transport protein family] |
-1.420 |
0.097 |
| 502 |
253689_at |
AT4G29770
|
unknown |
-1.418 |
1.329 |
| 503 |
258591_at |
AT3G04360
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-1.417 |
0.333 |
| 504 |
255174_at |
AT4G08030
|
[gypsy-like retrotransposon family (Athila), has a 1.5e-66 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-1.415 |
0.118 |
| 505 |
264051_at |
AT2G22340
|
unknown |
-1.415 |
0.139 |
| 506 |
260911_at |
AT1G02490
|
unknown |
-1.414 |
0.091 |
| 507 |
245673_at |
AT1G56690
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-1.413 |
0.053 |
| 508 |
249649_at |
AT5G37060
|
ATCHX24, ARABIDOPSIS THALIANA CATION/H+ EXCHANGER 24, CHX24, cation/H+ exchanger 24 |
-1.412 |
0.030 |
| 509 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-1.410 |
0.137 |
| 510 |
263055_at |
AT2G04600
|
unknown |
-1.409 |
0.193 |
| 511 |
253878_at |
AT4G27550
|
ATTPS4, ARABIDOPSIS THALIANA TREHALOSE PHOSPHATASE/SYNTHASE 4, TPS4, trehalose-6-phosphatase synthase S4 |
-1.409 |
0.071 |
| 512 |
257123_at |
AT3G20030
|
[F-box and associated interaction domains-containing protein] |
-1.407 |
0.105 |
| 513 |
248791_at |
AT5G47350
|
[alpha/beta-Hydrolases superfamily protein] |
-1.407 |
0.070 |
| 514 |
248585_at |
AT5G49640
|
unknown |
-1.407 |
0.080 |
| 515 |
257259_at |
AT3G22090
|
unknown |
-1.406 |
0.896 |
| 516 |
250759_at |
AT5G06020
|
[Plant self-incompatibility protein S1 family] |
-1.402 |
0.139 |
| 517 |
266723_at |
AT2G03190
|
ASK16, SKP1-like 16, SK16, SKP1-like 16 |
-1.402 |
0.120 |
| 518 |
252846_at |
AT3G42160
|
[Pectin lyase-like superfamily protein] |
-1.402 |
0.102 |
| 519 |
247142_at |
AT5G65570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-1.402 |
0.310 |
| 520 |
245549_at |
AT4G15320
|
ATCSLB06, cellulose synthase-like B6, ATCSLB6, CELLULOSE SYNTHASE LIKE B6, CSLB06, CSLB06, cellulose synthase-like B6 |
-1.402 |
0.183 |
| 521 |
260634_at |
AT1G62410
|
[MIF4G domain-containing protein] |
-1.402 |
0.127 |
| 522 |
262735_at |
AT1G28630
|
unknown |
-1.399 |
0.238 |
| 523 |
258960_at |
AT3G10590
|
[Duplicated homeodomain-like superfamily protein] |
-1.398 |
0.172 |
| 524 |
255746_at |
AT1G32020
|
[F-box family protein] |
-1.398 |
0.203 |
| 525 |
251294_at |
AT3G61940
|
ATMTPA1, MTPA1 |
-1.397 |
0.056 |
| 526 |
252857_at |
AT4G39756
|
[Galactose oxidase/kelch repeat superfamily protein] |
-1.397 |
0.176 |
| 527 |
265859_at |
AT2G01700
|
unknown |
-1.397 |
0.134 |
| 528 |
255262_at |
AT4G05140
|
[Nucleoside transporter family protein] |
-1.396 |
0.105 |
| 529 |
259798_at |
AT1G64310
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-1.395 |
0.165 |
| 530 |
258464_at |
AT3G17360
|
POK1, phragmoplast orienting kinesin 1 |
-1.394 |
0.116 |
| 531 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-1.393 |
0.024 |
| 532 |
262332_at |
AT1G64030
|
ATSRP3, SRP3, serpin 3 |
-1.392 |
0.150 |
| 533 |
258065_at |
AT3G25960
|
[Pyruvate kinase family protein] |
-1.392 |
0.179 |
| 534 |
266637_at |
AT2G35600
|
ATBRXL1, ARABIDOPSIS THALIANA BREVIS RADIX LIKE 1, BRXL1, BREVIS RADIX-like 1 |
-1.390 |
0.004 |
| 535 |
264235_at |
AT1G54560
|
ATXIE, MYOSIN XI E, XIE |
-1.390 |
0.043 |
| 536 |
245257_at |
AT4G14640
|
AtCML8, calmodulin-like 8, CAM8, calmodulin 8, CML8 |
-1.390 |
-0.018 |
| 537 |
264768_at |
AT1G61410
|
[DNA double-strand break repair and VJ recombination XRCC4] |
-1.389 |
0.141 |
| 538 |
248316_at |
AT5G52670
|
[Copper transport protein family] |
-1.387 |
-0.036 |
| 539 |
245503_at |
AT4G15650
|
unknown |
-1.386 |
0.101 |
| 540 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
-1.386 |
-0.098 |
| 541 |
259777_at |
AT1G29570
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
-1.385 |
0.203 |
| 542 |
257520_at |
AT3G07710
|
unknown |
-1.383 |
0.210 |
| 543 |
259425_at |
AT1G01460
|
ATPIPK11, PIPK11 |
-1.382 |
0.114 |
| 544 |
252539_at |
AT3G45730
|
unknown |
-1.382 |
0.279 |
| 545 |
263244_at |
AT2G31480
|
unknown |
-1.381 |
0.117 |
| 546 |
263309_at |
AT2G10370
|
unknown |
-1.380 |
0.069 |
| 547 |
255182_at |
AT4G08130
|
unknown |
-1.380 |
0.277 |
| 548 |
257570_at |
AT3G13662
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-1.379 |
0.163 |
| 549 |
249880_at |
AT5G23180
|
unknown |
-1.378 |
0.089 |
| 550 |
263062_at |
AT2G18180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-1.378 |
0.244 |
| 551 |
264143_at |
AT1G79330
|
AMC6, ATMC5, metacaspase 5, ATMCP2b, metacaspase 2b, MC5, metacaspase 5, MCP2b, metacaspase 2b |
-1.378 |
0.098 |
| 552 |
256477_at |
AT1G42690
|
[pseudogene] |
-1.376 |
0.585 |
| 553 |
257167_at |
AT3G24340
|
CHR40, chromatin remodeling 40 |
-1.376 |
0.599 |
| 554 |
247343_at |
AT5G63740
|
[RING/U-box superfamily protein] |
-1.376 |
0.070 |
| 555 |
255002_at |
AT4G09940
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.376 |
0.208 |
| 556 |
255415_at |
AT4G03160
|
unknown |
-1.375 |
0.137 |
| 557 |
251950_at |
AT3G53600
|
[C2H2-type zinc finger family protein] |
-1.373 |
0.193 |
| 558 |
265351_at |
AT2G22630
|
AGL17, AGAMOUS-like 17 |
-1.371 |
0.085 |
| 559 |
261245_at |
AT1G20135
|
[GDSL-like Lipase/Acylhydrolase family protein] |
-1.371 |
0.105 |
| 560 |
250242_at |
AT5G13620
|
unknown |
-1.370 |
0.205 |
| 561 |
263006_at |
AT1G54240
|
[winged-helix DNA-binding transcription factor family protein] |
-1.369 |
0.198 |
| 562 |
266249_at |
AT2G27630
|
[Ubiquitin carboxyl-terminal hydrolase-related protein] |
-1.367 |
0.130 |
| 563 |
254595_at |
AT4G18960
|
AG, AGAMOUS |
-1.367 |
0.108 |
| 564 |
261281_at |
AT1G35740
|
[pseudogene] |
-1.367 |
0.015 |
| 565 |
264276_at |
AT1G60380
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
-1.366 |
0.149 |
| 566 |
252191_at |
AT3G50180
|
unknown |
-1.366 |
0.045 |
| 567 |
265489_at |
AT2G15640
|
[F-box family protein] |
-1.365 |
0.126 |
| 568 |
265506_at |
AT2G15540
|
[non-LTR retrotransposon family (LINE), has a 2.7e-33 P-value blast match to GB:NP_038607 L1 repeat, Tf subfamily, member 9 (LINE-element) (Mus musculus)] |
-1.365 |
0.098 |
| 569 |
253472_at |
AT4G32230
|
unknown |
-1.363 |
0.149 |
| 570 |
251085_at |
AT5G01440
|
unknown |
-1.363 |
0.115 |
| 571 |
248929_at |
AT5G46000
|
[Mannose-binding lectin superfamily protein] |
-1.362 |
0.088 |
| 572 |
250416_at |
AT5G11220
|
unknown |
-1.361 |
0.072 |
| 573 |
257640_at |
AT3G25750
|
unknown |
-1.359 |
0.173 |
| 574 |
263392_at |
AT2G10490
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.0e-43 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-1.359 |
0.056 |
| 575 |
262857_at |
AT1G14930
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.359 |
-0.045 |
| 576 |
247798_at |
AT5G58830
|
[Subtilisin-like serine endopeptidase family protein] |
-1.359 |
0.063 |
| 577 |
254139_at |
AT4G24600
|
unknown |
-1.359 |
0.175 |
| 578 |
257446_at |
AT2G10440
|
unknown |
-1.358 |
0.090 |
| 579 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
-1.358 |
1.727 |
| 580 |
250579_at |
AT5G07930
|
MCT2, MEI2 C-terminal RRM only like 2 |
-1.358 |
0.089 |
| 581 |
250166_at |
AT5G15300
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-1.357 |
0.063 |
| 582 |
255075_at |
AT4G09110
|
[RING/U-box superfamily protein] |
-1.356 |
0.130 |
| 583 |
249226_at |
AT5G42170
|
[SGNH hydrolase-type esterase superfamily protein] |
-1.356 |
0.101 |
| 584 |
257563_at |
AT3G19610
|
unknown |
-1.356 |
0.165 |
| 585 |
253910_at |
AT4G27290
|
[S-locus lectin protein kinase family protein] |
-1.355 |
0.147 |
| 586 |
264499_at |
AT1G30795
|
[Glycine-rich protein family] |
-1.355 |
0.116 |
| 587 |
258290_at |
AT3G23460
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.355 |
0.063 |
| 588 |
250574_at |
AT5G08230
|
[Tudor/PWWP/MBT domain-containing protein] |
-1.354 |
0.045 |
| 589 |
255315_at |
AT4G04165
|
[pseudogene] |
-1.354 |
0.179 |
| 590 |
265401_at |
AT2G10970
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.353 |
0.060 |
| 591 |
265980_at |
AT2G11165
|
[pseudogene] |
-1.353 |
0.191 |
| 592 |
250959_at |
AT5G02920
|
[F-box/RNI-like superfamily protein] |
-1.353 |
0.232 |
| 593 |
258940_at |
AT3G09930
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.352 |
0.119 |
| 594 |
256039_at |
AT1G19190
|
[alpha/beta-Hydrolases superfamily protein] |
-1.351 |
0.029 |
| 595 |
246569_at |
AT5G14980
|
[alpha/beta-Hydrolases superfamily protein] |
-1.350 |
0.071 |
| 596 |
259370_at |
AT1G69050
|
unknown |
-1.350 |
0.075 |
| 597 |
246706_at |
AT5G28130
|
unknown |
-1.350 |
0.045 |
| 598 |
247711_at |
AT5G59270
|
LecRK-II.2, L-type lectin receptor kinase II.2 |
-1.348 |
0.084 |
| 599 |
263454_at |
AT2G22160
|
[Cysteine proteinases superfamily protein] |
-1.348 |
0.066 |
| 600 |
256391_at |
AT3G06090
|
unknown |
-1.348 |
0.134 |