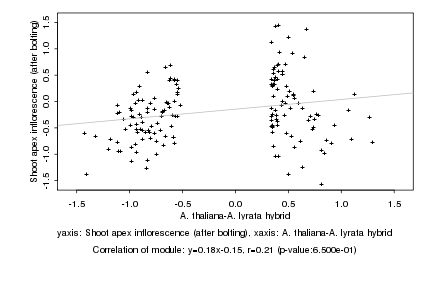
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
A. thaliana-A. lyrata hybrid |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
1.294 |
-0.774 |
| 2 |
256460_at |
AT1G36240
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
1.264 |
-0.303 |
| 3 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
1.118 |
0.135 |
| 4 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
1.092 |
-0.702 |
| 5 |
250773_at |
AT5G05430
|
[RNA-binding protein] |
1.059 |
-0.161 |
| 6 |
258370_at |
AT3G14395
|
unknown |
0.933 |
-0.447 |
| 7 |
254699_at |
AT4G17990
|
unknown |
0.907 |
-0.783 |
| 8 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.852 |
-0.729 |
| 9 |
259054_at |
AT3G03480
|
CHAT, acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase |
0.833 |
-0.972 |
| 10 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.811 |
-0.926 |
| 11 |
260934_at |
AT1G45100
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.804 |
-1.556 |
| 12 |
255576_at |
AT4G01440
|
UMAMIT31, Usually multiple acids move in and out Transporters 31 |
0.780 |
-0.261 |
| 13 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
0.765 |
-0.243 |
| 14 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
0.744 |
-0.330 |
| 15 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.736 |
0.192 |
| 16 |
267569_at |
AT2G30790
|
PSBP-2, photosystem II subunit P-2 |
0.732 |
-0.488 |
| 17 |
260930_at |
AT1G02620
|
[Ras-related small GTP-binding family protein] |
0.725 |
-0.521 |
| 18 |
252677_at |
AT3G44320
|
AtNIT3, NITRILASE 3, NIT3, nitrilase 3 |
0.701 |
-0.272 |
| 19 |
245814_at |
AT1G49910
|
BUB3.2, BUB (BUDDING UNINHIBITED BY BENZYMIDAZOL) 3.2 |
0.685 |
-0.359 |
| 20 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
0.665 |
1.367 |
| 21 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
0.647 |
0.850 |
| 22 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.631 |
-1.237 |
| 23 |
248691_at |
AT5G48310
|
unknown |
0.629 |
-0.125 |
| 24 |
264181_at |
AT1G65350
|
UBQ13, ubiquitin 13 |
0.613 |
-0.741 |
| 25 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
0.592 |
-0.027 |
| 26 |
257473_at |
AT1G33840
|
unknown |
0.550 |
-0.863 |
| 27 |
253729_at |
AT4G29360
|
[O-Glycosyl hydrolases family 17 protein] |
0.549 |
0.061 |
| 28 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
0.547 |
0.125 |
| 29 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.543 |
0.139 |
| 30 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
0.538 |
0.928 |
| 31 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.524 |
-0.662 |
| 32 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.519 |
-0.130 |
| 33 |
260151_at |
AT1G52910
|
unknown |
0.506 |
0.204 |
| 34 |
250783_at |
AT5G05260
|
CYP79A2, cytochrome p450 79a2 |
0.500 |
-1.372 |
| 35 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
0.495 |
1.216 |
| 36 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
0.490 |
0.096 |
| 37 |
263151_at |
AT1G54120
|
unknown |
0.477 |
-0.588 |
| 38 |
248074_at |
AT5G55730
|
FLA1, FASCICLIN-like arabinogalactan 1 |
0.475 |
0.296 |
| 39 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.471 |
-0.031 |
| 40 |
246231_at |
AT4G37080
|
unknown |
0.469 |
0.708 |
| 41 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
0.457 |
-0.252 |
| 42 |
256832_at |
AT3G22880
|
ARLIM15, ARABIDOPSIS HOMOLOG OF LILY MESSAGES INDUCED AT MEIOSIS 15, ATDMC1, ARABIDOPSIS THALIANA DISRUPTION OF MEIOTIC CONTROL 1, DMC1, DISRUPTION OF MEIOTIC CONTROL 1 |
0.442 |
0.515 |
| 43 |
265057_at |
AT1G52140
|
unknown |
0.441 |
0.581 |
| 44 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.440 |
0.008 |
| 45 |
258122_at |
AT3G14570
|
ATGSL04, glucan synthase-like 4, atgsl4, gsl04, GSL04, glucan synthase-like 4 |
0.440 |
0.515 |
| 46 |
266891_at |
AT2G44690
|
ARAC9, Arabidopsis RAC-like 9, ATROP8, RHO-RELATED PROTEIN FROM PLANTS 8, ROP8, RHO-RELATED PROTEIN FROM PLANTS 8 |
0.427 |
-0.072 |
| 47 |
247625_at |
AT5G60200
|
TMO6, TARGET OF MONOPTEROS 6 |
0.410 |
0.935 |
| 48 |
261129_at |
AT1G04820
|
TOR2, TORTIFOLIA 2, TUA4, tubulin alpha-4 chain |
0.400 |
1.456 |
| 49 |
266913_at |
AT2G45890
|
ATROPGEF4, RHO GUANYL-NUCLEOTIDE EXCHANGE FACTOR 4, GEF4, guanine exchange factor 4, RHS11, ROOT HAIR SPECIFIC 11, ROPGEF4, ROP (rho of plants) guanine nucleotide exchange factor 4 |
0.400 |
-1.032 |
| 50 |
253891_at |
AT4G27720
|
[Major facilitator superfamily protein] |
0.399 |
0.708 |
| 51 |
258241_at |
AT3G27650
|
LBD25, LOB domain-containing protein 25 |
0.398 |
0.579 |
| 52 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
0.397 |
-0.449 |
| 53 |
261025_at |
AT1G01225
|
[NC domain-containing protein-related] |
0.396 |
0.691 |
| 54 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
0.394 |
-0.361 |
| 55 |
251961_at |
AT3G53620
|
AtPPa4, pyrophosphorylase 4, PPa4, pyrophosphorylase 4 |
0.391 |
0.230 |
| 56 |
253010_at |
AT4G37750
|
ANT, AINTEGUMENTA, CKC, CKC1, COMPLEMENTING A PROTEIN KINASE C MUTANT 1, DRG, DRAGON |
0.390 |
0.421 |
| 57 |
248924_at |
AT5G45960
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.384 |
-0.263 |
| 58 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.383 |
-0.337 |
| 59 |
267402_at |
AT2G26180
|
IQD6, IQ-domain 6 |
0.378 |
1.436 |
| 60 |
246512_at |
AT5G15630
|
COBL4, COBRA-LIKE4, IRX6, IRREGULAR XYLEM 6 |
0.377 |
-1.032 |
| 61 |
245113_at |
AT2G41660
|
MIZ1, mizu-kussei 1 |
0.374 |
0.465 |
| 62 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
0.372 |
-0.168 |
| 63 |
247441_at |
AT5G62710
|
[Leucine-rich repeat protein kinase family protein] |
0.368 |
0.650 |
| 64 |
248257_at |
AT5G53410
|
unknown |
0.364 |
0.417 |
| 65 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
0.364 |
0.409 |
| 66 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
0.362 |
0.332 |
| 67 |
251547_at |
AT3G58860
|
[F-box/RNI-like superfamily protein] |
0.361 |
0.415 |
| 68 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.359 |
0.340 |
| 69 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.358 |
0.114 |
| 70 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
0.356 |
0.542 |
| 71 |
255417_at |
AT4G03190
|
AFB1, AUXIN SIGNALING F BOX PROTEIN 1, ATGRH1, GRH1, GRR1-like protein 1 |
0.356 |
0.619 |
| 72 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
0.351 |
-0.852 |
| 73 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
0.350 |
-0.452 |
| 74 |
257247_at |
AT3G24140
|
FMA, FAMA |
0.346 |
-0.470 |
| 75 |
248457_at |
AT5G51420
|
[long-chain-alcohol O-fatty-acyltransferase family protein / wax synthase family protein] |
0.345 |
0.304 |
| 76 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
0.344 |
-0.474 |
| 77 |
257342_at |
AT1G42365
|
[gypsy-like retrotransposon family (Athila), has a 9.0e-234 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.344 |
-0.231 |
| 78 |
257647_at |
AT3G25805
|
unknown |
0.342 |
0.309 |
| 79 |
257480_at |
AT1G15640
|
unknown |
0.341 |
-0.571 |
| 80 |
256704_at |
AT3G30280
|
[HXXXD-type acyl-transferase family protein] |
0.339 |
-0.117 |
| 81 |
267129_at |
AT2G23380
|
CLF, CURLY LEAF, ICU1, INCURVATA 1, SDG1, SETDOMAIN GROUP 1, SET1, SETDOMAIN 1 |
0.338 |
0.328 |
| 82 |
255577_at |
AT4G01410
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.335 |
-0.354 |
| 83 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.335 |
-0.466 |
| 84 |
266905_at |
AT2G34560
|
AtCCP1, CCP1, conserved in ciliated species and in the land plants 1 |
0.333 |
0.436 |
| 85 |
245524_at |
AT4G15920
|
AtSWEET17, SWEET17 |
0.333 |
1.135 |
| 86 |
253533_at |
AT4G31590
|
ATCSLC05, CELLULOSE-SYNTHASE LIKE C5, ATCSLC5, Cellulose-synthase-like C5, CSLC05, CELLULOSE-SYNTHASE LIKE C5, CSLC5, Cellulose-synthase-like C5 |
0.332 |
-0.283 |
| 87 |
256442_at |
AT3G10930
|
unknown |
-1.428 |
-0.602 |
| 88 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-1.414 |
-1.379 |
| 89 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
-1.324 |
-0.656 |
| 90 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-1.200 |
-0.909 |
| 91 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
-1.190 |
-0.718 |
| 92 |
263182_at |
AT1G05575
|
unknown |
-1.123 |
-0.066 |
| 93 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
-1.122 |
-0.771 |
| 94 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
-1.119 |
-0.210 |
| 95 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
-1.109 |
-0.935 |
| 96 |
260005_at |
AT1G67920
|
unknown |
-1.096 |
-0.192 |
| 97 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
-1.091 |
-0.941 |
| 98 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
-1.062 |
-0.333 |
| 99 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-1.039 |
-0.518 |
| 100 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-1.001 |
-0.442 |
| 101 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
-0.995 |
-0.124 |
| 102 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.988 |
-0.863 |
| 103 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.986 |
-0.167 |
| 104 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
-0.984 |
-1.135 |
| 105 |
258203_at |
AT3G13950
|
unknown |
-0.984 |
-0.269 |
| 106 |
247800_at |
AT5G58570
|
unknown |
-0.972 |
0.147 |
| 107 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
-0.971 |
-0.288 |
| 108 |
259479_at |
AT1G19020
|
unknown |
-0.953 |
-0.031 |
| 109 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.946 |
-0.805 |
| 110 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-0.942 |
0.185 |
| 111 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.940 |
-0.530 |
| 112 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.937 |
-0.436 |
| 113 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.935 |
-0.966 |
| 114 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
-0.928 |
-0.570 |
| 115 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.918 |
0.033 |
| 116 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
-0.912 |
-0.232 |
| 117 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-0.911 |
0.303 |
| 118 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.902 |
-0.522 |
| 119 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
-0.895 |
-0.287 |
| 120 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.885 |
-0.388 |
| 121 |
266658_at |
AT2G25735
|
unknown |
-0.884 |
-0.540 |
| 122 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-0.883 |
0.036 |
| 123 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
-0.882 |
-0.708 |
| 124 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.855 |
-0.587 |
| 125 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
-0.846 |
-1.267 |
| 126 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.840 |
-0.196 |
| 127 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
-0.839 |
-1.111 |
| 128 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
-0.835 |
0.556 |
| 129 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
-0.831 |
-0.121 |
| 130 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-0.826 |
-0.532 |
| 131 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.821 |
-0.584 |
| 132 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-0.806 |
-0.029 |
| 133 |
262615_at |
AT1G13950
|
ATELF5A-1, EUKARYOTIC ELONGATION FACTOR 5A-1, EIF-5A, EIF5A, EUKARYOTIC ELONGATION FACTOR 5A, ELF5A-1, eukaryotic elongation factor 5A-1 |
-0.804 |
-0.696 |
| 134 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.796 |
-0.456 |
| 135 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.770 |
-0.590 |
| 136 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
-0.769 |
-0.142 |
| 137 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
-0.767 |
0.059 |
| 138 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
-0.753 |
-1.005 |
| 139 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-0.750 |
-0.744 |
| 140 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.738 |
-0.413 |
| 141 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
-0.714 |
-0.540 |
| 142 |
257591_at |
AT3G24900
|
AtRLP39, receptor like protein 39, RLP39, receptor like protein 39 |
-0.706 |
-0.279 |
| 143 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.698 |
-0.184 |
| 144 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
-0.692 |
-0.200 |
| 145 |
252345_at |
AT3G48640
|
unknown |
-0.687 |
-0.831 |
| 146 |
266017_at |
AT2G18690
|
unknown |
-0.677 |
-0.163 |
| 147 |
266780_at |
AT2G29110
|
ATGLR2.8, glutamate receptor 2.8, GLR2.8, GLR2.8, glutamate receptor 2.8 |
-0.668 |
-0.654 |
| 148 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
-0.663 |
0.649 |
| 149 |
246842_at |
AT5G26731
|
unknown |
-0.653 |
-0.009 |
| 150 |
252681_at |
AT3G44350
|
anac061, NAC domain containing protein 61, NAC061, NAC domain containing protein 61 |
-0.646 |
-0.034 |
| 151 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.641 |
-0.034 |
| 152 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.629 |
-0.110 |
| 153 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.628 |
0.402 |
| 154 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.621 |
0.439 |
| 155 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.615 |
0.694 |
| 156 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
-0.608 |
-0.470 |
| 157 |
250796_at |
AT5G05300
|
unknown |
-0.599 |
-0.255 |
| 158 |
246144_at |
AT5G20110
|
[Dynein light chain type 1 family protein] |
-0.594 |
-0.674 |
| 159 |
245375_at |
AT4G17660
|
[Protein kinase superfamily protein] |
-0.587 |
-0.667 |
| 160 |
247047_at |
AT5G66650
|
unknown |
-0.580 |
-0.786 |
| 161 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
-0.579 |
0.414 |
| 162 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
-0.577 |
0.011 |
| 163 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.577 |
0.435 |
| 164 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.574 |
-0.281 |
| 165 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
-0.563 |
0.335 |
| 166 |
255596_at |
AT4G01720
|
AtWRKY47, WRKY47 |
-0.554 |
0.189 |
| 167 |
265327_at |
AT2G18210
|
unknown |
-0.552 |
-0.269 |
| 168 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
-0.549 |
0.414 |
| 169 |
249622_at |
AT5G37550
|
unknown |
-0.549 |
0.145 |
| 170 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
-0.542 |
0.252 |
| 171 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
-0.527 |
-0.062 |