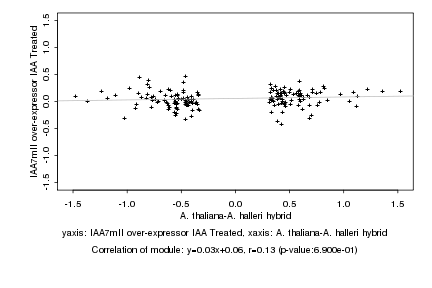
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
A. thaliana-A. halleri hybrid |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
255471_at |
AT4G03050
|
AOP3 |
1.521 |
0.198 |
| 2 |
256096_at |
AT1G13650
|
unknown |
1.358 |
0.193 |
| 3 |
256460_at |
AT1G36240
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
1.211 |
0.237 |
| 4 |
253658_at |
AT4G30120
|
ATHMA3, A. THALIANA HEAVY METAL ATPASE 3, HMA3, heavy metal atpase 3 |
1.122 |
0.106 |
| 5 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.116 |
-0.076 |
| 6 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
1.090 |
0.177 |
| 7 |
250773_at |
AT5G05430
|
[RNA-binding protein] |
1.046 |
0.007 |
| 8 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
0.969 |
0.131 |
| 9 |
267569_at |
AT2G30790
|
PSBP-2, photosystem II subunit P-2 |
0.843 |
0.036 |
| 10 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.818 |
0.244 |
| 11 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.811 |
0.290 |
| 12 |
251949_at |
AT3G53590
|
[Leucine-rich repeat protein kinase family protein] |
0.781 |
0.181 |
| 13 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
0.772 |
-0.001 |
| 14 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
0.756 |
-0.065 |
| 15 |
249461_at |
AT5G39640
|
[Putative endonuclease or glycosyl hydrolase] |
0.742 |
0.164 |
| 16 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.704 |
0.229 |
| 17 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.703 |
0.182 |
| 18 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.699 |
-0.241 |
| 19 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
0.678 |
-0.063 |
| 20 |
262416_at |
AT1G49390
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.676 |
-0.307 |
| 21 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.676 |
0.091 |
| 22 |
253754_at |
AT4G29020
|
[glycine-rich protein] |
0.659 |
0.112 |
| 23 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
0.622 |
0.091 |
| 24 |
249004_at |
AT5G44570
|
unknown |
0.621 |
0.042 |
| 25 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
0.616 |
-0.133 |
| 26 |
250135_at |
AT5G15360
|
unknown |
0.606 |
0.118 |
| 27 |
260930_at |
AT1G02620
|
[Ras-related small GTP-binding family protein] |
0.599 |
0.149 |
| 28 |
262464_at |
AT1G50280
|
[Phototropic-responsive NPH3 family protein] |
0.595 |
0.011 |
| 29 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.590 |
0.378 |
| 30 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.590 |
0.126 |
| 31 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.588 |
0.041 |
| 32 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
0.587 |
0.210 |
| 33 |
253532_at |
AT4G31570
|
unknown |
0.587 |
0.102 |
| 34 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.580 |
0.114 |
| 35 |
248734_at |
AT5G48090
|
ELP1, EDM2-like protein1 |
0.576 |
-0.058 |
| 36 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
0.574 |
0.193 |
| 37 |
259522_at |
AT1G12490
|
[F-box associated ubiquitination effector family protein] |
0.556 |
0.153 |
| 38 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
0.529 |
0.148 |
| 39 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
0.511 |
0.019 |
| 40 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
0.504 |
-0.052 |
| 41 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.501 |
0.228 |
| 42 |
253050_at |
AT4G37450
|
AGP18, arabinogalactan protein 18, ATAGP18 |
0.493 |
0.174 |
| 43 |
257275_at |
AT3G14450
|
CID9, CTC-interacting domain 9 |
0.466 |
0.130 |
| 44 |
246817_at |
AT5G27240
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.462 |
-0.012 |
| 45 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.458 |
-0.079 |
| 46 |
254566_at |
AT4G19240
|
unknown |
0.453 |
0.163 |
| 47 |
262040_at |
AT1G80080
|
AtRLP17, Receptor Like Protein 17, TMM, TOO MANY MOUTHS |
0.452 |
0.177 |
| 48 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
0.452 |
-0.053 |
| 49 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
0.448 |
0.266 |
| 50 |
257417_at |
AT1G10110
|
[F-box family protein] |
0.444 |
-0.035 |
| 51 |
252345_at |
AT3G48640
|
unknown |
0.443 |
0.204 |
| 52 |
266718_at |
AT2G46800
|
ATCDF1, A. THALIANA CATION DIFFUSION FACILITATOR 1, ATMTP1, MTP1, metal tolerance protein 1, OZS1, OVERLY ZINC SENSITIVE 1, ZAT, zinc transporter of Arabidopsis thaliana, ZAT1, ZINC TRANSPORTER OF ARABIDOPSIS THALIANA 1 |
0.430 |
0.004 |
| 53 |
255613_at |
AT4G01270
|
[RING/U-box superfamily protein] |
0.429 |
0.049 |
| 54 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
0.426 |
-0.200 |
| 55 |
263365_at |
AT2G20550
|
[HSP40/DnaJ peptide-binding protein] |
0.423 |
-0.031 |
| 56 |
260151_at |
AT1G52910
|
unknown |
0.422 |
0.162 |
| 57 |
258356_at |
AT3G14340
|
unknown |
0.422 |
-0.416 |
| 58 |
265057_at |
AT1G52140
|
unknown |
0.416 |
0.155 |
| 59 |
259912_at |
AT1G72670
|
iqd8, IQ-domain 8 |
0.415 |
0.095 |
| 60 |
267266_at |
AT2G23150
|
ATNRAMP3, NRAMP3, natural resistance-associated macrophage protein 3 |
0.413 |
0.124 |
| 61 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.412 |
0.233 |
| 62 |
245454_at |
AT4G16920
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.407 |
0.073 |
| 63 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
0.407 |
0.130 |
| 64 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
0.395 |
0.088 |
| 65 |
245929_at |
AT5G24760
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.394 |
-0.044 |
| 66 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.391 |
0.163 |
| 67 |
267019_at |
AT2G39130
|
[Transmembrane amino acid transporter family protein] |
0.385 |
-0.359 |
| 68 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
0.384 |
0.160 |
| 69 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.383 |
0.052 |
| 70 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.379 |
0.097 |
| 71 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
0.377 |
0.214 |
| 72 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
0.364 |
0.283 |
| 73 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.357 |
-0.072 |
| 74 |
253729_at |
AT4G29360
|
[O-Glycosyl hydrolases family 17 protein] |
0.349 |
0.219 |
| 75 |
265587_at |
AT2G19980
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.343 |
0.003 |
| 76 |
264852_at |
AT2G17480
|
ATMLO8, MILDEW RESISTANCE LOCUS O 8, MLO8, MILDEW RESISTANCE LOCUS O 8 |
0.341 |
0.071 |
| 77 |
264235_at |
AT1G54560
|
ATXIE, MYOSIN XI E, XIE |
0.335 |
0.037 |
| 78 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.332 |
0.081 |
| 79 |
265425_at |
AT2G20770
|
GCL2, GCR2-like 2 |
0.330 |
0.036 |
| 80 |
261685_at |
AT1G47290
|
3BETAHSD/D1, 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 1, AT3BETAHSD/D1, 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 1 |
0.328 |
-0.196 |
| 81 |
264104_at |
AT2G13750
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.5e-17 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.326 |
0.068 |
| 82 |
265382_at |
AT2G16790
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.324 |
0.091 |
| 83 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
0.324 |
0.244 |
| 84 |
250133_at |
AT5G16400
|
ATF2, TRXF2, thioredoxin F2 |
0.320 |
0.175 |
| 85 |
263688_at |
AT1G26920
|
unknown |
0.319 |
0.316 |
| 86 |
262920_at |
AT1G79500
|
AtkdsA1 |
0.317 |
0.054 |
| 87 |
245574_at |
AT4G14750
|
IQD19, IQ-domain 19 |
0.314 |
-0.012 |
| 88 |
265611_at |
AT2G25510
|
unknown |
-1.480 |
0.096 |
| 89 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-1.368 |
0.005 |
| 90 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-1.242 |
0.186 |
| 91 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.182 |
0.067 |
| 92 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-1.109 |
0.127 |
| 93 |
258027_at |
AT3G19515
|
unknown |
-1.027 |
-0.305 |
| 94 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
-0.980 |
0.251 |
| 95 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.925 |
-0.117 |
| 96 |
254894_at |
AT4G11840
|
PLDGAMMA3, phospholipase D gamma 3 |
-0.921 |
-0.053 |
| 97 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.899 |
0.150 |
| 98 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
-0.887 |
0.460 |
| 99 |
254481_at |
AT4G20480
|
[Putative endonuclease or glycosyl hydrolase] |
-0.868 |
0.089 |
| 100 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.822 |
0.057 |
| 101 |
245451_at |
AT4G16890
|
BAL, BALL, SNC1, SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 |
-0.816 |
0.143 |
| 102 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.814 |
0.332 |
| 103 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.812 |
0.404 |
| 104 |
256617_at |
AT3G22240
|
unknown |
-0.795 |
0.264 |
| 105 |
262615_at |
AT1G13950
|
ATELF5A-1, EUKARYOTIC ELONGATION FACTOR 5A-1, EIF-5A, EIF5A, EUKARYOTIC ELONGATION FACTOR 5A, ELF5A-1, eukaryotic elongation factor 5A-1 |
-0.783 |
-0.102 |
| 106 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.780 |
0.084 |
| 107 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
-0.774 |
0.034 |
| 108 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.766 |
0.101 |
| 109 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.744 |
0.044 |
| 110 |
255627_at |
AT4G00955
|
unknown |
-0.720 |
-0.004 |
| 111 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
-0.717 |
0.014 |
| 112 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
-0.696 |
0.204 |
| 113 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
-0.656 |
0.034 |
| 114 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
-0.651 |
0.130 |
| 115 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
-0.646 |
-0.003 |
| 116 |
262669_at |
AT1G62850
|
[Class I peptide chain release factor] |
-0.631 |
-0.034 |
| 117 |
257741_at |
AT3G27340
|
unknown |
-0.621 |
-0.142 |
| 118 |
252659_at |
AT3G44430
|
unknown |
-0.621 |
0.237 |
| 119 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
-0.620 |
-0.064 |
| 120 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.614 |
-0.105 |
| 121 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.606 |
0.209 |
| 122 |
258228_at |
AT3G27610
|
[Nucleotidylyl transferase superfamily protein] |
-0.596 |
0.100 |
| 123 |
252165_at |
AT3G50550
|
unknown |
-0.568 |
-0.196 |
| 124 |
260005_at |
AT1G67920
|
unknown |
-0.567 |
0.052 |
| 125 |
248079_at |
AT5G55790
|
unknown |
-0.565 |
-0.031 |
| 126 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
-0.562 |
-0.015 |
| 127 |
253422_at |
AT4G32240
|
unknown |
-0.559 |
0.017 |
| 128 |
259529_at |
AT1G12400
|
[Nucleotide excision repair, TFIIH, subunit TTDA] |
-0.555 |
-0.244 |
| 129 |
247988_at |
AT5G56910
|
[Proteinase inhibitor I25, cystatin, conserved region] |
-0.555 |
0.123 |
| 130 |
265245_at |
AT2G43060
|
AtIBH1, IBH1, ILI1 binding bHLH 1 |
-0.553 |
-0.101 |
| 131 |
254586_at |
AT4G19510
|
[Disease resistance protein (TIR-NBS-LRR class)] |
-0.551 |
-0.219 |
| 132 |
245384_at |
AT4G16790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.551 |
-0.115 |
| 133 |
250931_at |
AT5G03200
|
LUL1, LOG2-LIKE UBIQUITIN LIGASE1 |
-0.551 |
-0.040 |
| 134 |
252593_at |
AT3G45590
|
ATSEN1, splicing endonuclease 1, SEN1, splicing endonuclease 1 |
-0.544 |
-0.031 |
| 135 |
251346_at |
AT3G60980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.543 |
0.148 |
| 136 |
245055_at |
AT2G26470
|
unknown |
-0.543 |
-0.134 |
| 137 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
-0.538 |
0.123 |
| 138 |
256568_at |
AT3G19520
|
unknown |
-0.533 |
0.044 |
| 139 |
256303_at |
AT1G69550
|
[disease resistance protein (TIR-NBS-LRR class)] |
-0.533 |
0.062 |
| 140 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
-0.506 |
0.079 |
| 141 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
-0.485 |
0.355 |
| 142 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.482 |
0.063 |
| 143 |
252282_at |
AT3G49360
|
PGL2, 6-phosphogluconolactonase 2 |
-0.481 |
0.181 |
| 144 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.481 |
0.206 |
| 145 |
251037_at |
AT5G02100
|
ORP3A, OSBP(OXYSTEROL BINDING PROTEIN)-RELATED PROTEIN 3A, UNE18, UNFERTILIZED EMBRYO SAC 18 |
-0.470 |
-0.046 |
| 146 |
260966_at |
AT1G12220
|
RPS5, RESISTANT TO P. SYRINGAE 5 |
-0.466 |
-0.319 |
| 147 |
247105_at |
AT5G66630
|
DAR5, DA1-related protein 5 |
-0.464 |
0.019 |
| 148 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
-0.463 |
-0.010 |
| 149 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.463 |
0.470 |
| 150 |
255260_at |
AT4G05040
|
[ankyrin repeat family protein] |
-0.460 |
-0.069 |
| 151 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
-0.455 |
0.001 |
| 152 |
249822_at |
AT5G23710
|
[DNA binding] |
-0.453 |
-0.021 |
| 153 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.446 |
-0.070 |
| 154 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.444 |
0.083 |
| 155 |
260110_at |
AT1G63350
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.438 |
0.045 |
| 156 |
264232_at |
AT1G67470
|
[Protein kinase superfamily protein] |
-0.438 |
-0.070 |
| 157 |
261317_at |
AT1G53030
|
[Cytochrome C oxidase copper chaperone (COX17)] |
-0.428 |
-0.000 |
| 158 |
266659_at |
AT2G25850
|
PAPS2, poly(A) polymerase 2 |
-0.415 |
0.050 |
| 159 |
248315_at |
AT5G52630
|
MEF1, mitochondrial RNA editing factor 1 |
-0.411 |
-0.265 |
| 160 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
-0.408 |
-0.001 |
| 161 |
259934_at |
AT1G71340
|
AtGDPD4, GDPD4, glycerophosphodiester phosphodiesterase 4 |
-0.408 |
0.015 |
| 162 |
266480_at |
AT2G31130
|
unknown |
-0.407 |
0.098 |
| 163 |
264658_at |
AT1G09910
|
[Rhamnogalacturonate lyase family protein] |
-0.404 |
-0.164 |
| 164 |
249838_at |
AT5G23460
|
unknown |
-0.400 |
-0.019 |
| 165 |
259479_at |
AT1G19020
|
unknown |
-0.394 |
0.038 |
| 166 |
261561_at |
AT1G01730
|
unknown |
-0.373 |
-0.030 |
| 167 |
248694_at |
AT5G48340
|
unknown |
-0.360 |
-0.016 |
| 168 |
247934_at |
AT5G57000
|
unknown |
-0.359 |
-0.055 |
| 169 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
-0.355 |
0.180 |
| 170 |
260189_at |
AT1G67550
|
URE, urease |
-0.347 |
-0.146 |
| 171 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
-0.346 |
0.128 |
| 172 |
247167_at |
AT5G65850
|
[F-box and associated interaction domains-containing protein] |
-0.346 |
0.134 |
| 173 |
262668_at |
AT1G62830
|
ATLSD1, ARABIDOPSIS LYSINE-SPECIFIC HISTONE DEMETHYLASE, ATSWP1, LDL1, LSD1-like 1, LSD1, LYSINE-SPECIFIC HISTONE DEMETHYLASE, SWP1 |
-0.340 |
-0.152 |