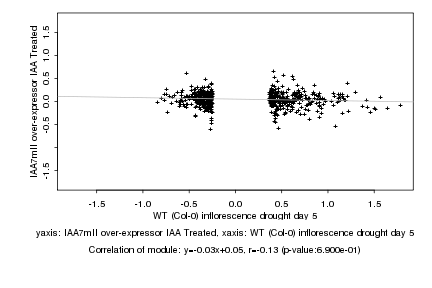
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT (Col-0) inflorescence drought day 5 |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
1.780 |
-0.080 |
| 2 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.642 |
-0.141 |
| 3 |
251988_at |
AT3G53300
|
CYP71B31, cytochrome P450, family 71, subfamily B, polypeptide 31 |
1.568 |
0.100 |
| 4 |
247061_at |
AT5G66780
|
unknown |
1.505 |
-0.159 |
| 5 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.499 |
-0.132 |
| 6 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.457 |
-0.230 |
| 7 |
263881_at |
AT2G21820
|
unknown |
1.422 |
-0.111 |
| 8 |
258327_at |
AT3G22640
|
PAP85 |
1.410 |
0.041 |
| 9 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
1.367 |
-0.091 |
| 10 |
246279_at |
AT4G36740
|
ATHB40, homeobox protein 40, HB-5, HB40, homeobox protein 40 |
1.291 |
0.210 |
| 11 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
1.212 |
-0.211 |
| 12 |
265983_at |
AT2G18550
|
ATHB21, homeobox protein 21, HB-2, homeobox-2, HB21, homeobox protein 21 |
1.208 |
0.392 |
| 13 |
262399_at |
AT1G49500
|
unknown |
1.204 |
0.121 |
| 14 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
1.185 |
0.037 |
| 15 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
1.169 |
0.023 |
| 16 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
1.159 |
0.068 |
| 17 |
256021_at |
AT1G58270
|
ZW9 |
1.153 |
-0.246 |
| 18 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
1.151 |
0.161 |
| 19 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.147 |
0.118 |
| 20 |
265387_at |
AT2G20670
|
unknown |
1.132 |
0.163 |
| 21 |
257186_at |
AT3G13130
|
unknown |
1.131 |
0.033 |
| 22 |
249117_at |
AT5G43840
|
AT-HSFA6A, heat shock transcription factor A6A, HSFA6A, heat shock transcription factor A6A |
1.123 |
0.092 |
| 23 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.114 |
0.005 |
| 24 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
1.113 |
0.073 |
| 25 |
264774_at |
AT1G22890
|
unknown |
1.106 |
0.166 |
| 26 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
1.101 |
-0.013 |
| 27 |
248969_at |
AT5G45310
|
unknown |
1.076 |
-0.525 |
| 28 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.063 |
0.120 |
| 29 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
1.055 |
-0.079 |
| 30 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
1.053 |
0.190 |
| 31 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
1.029 |
0.021 |
| 32 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.972 |
0.020 |
| 33 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.955 |
0.056 |
| 34 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.947 |
-0.045 |
| 35 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.938 |
0.048 |
| 36 |
253152_at |
AT4G35690
|
unknown |
0.936 |
0.073 |
| 37 |
257853_at |
AT3G12960
|
SMP1, seed maturation protein 1 |
0.924 |
-0.255 |
| 38 |
256114_at |
AT1G16850
|
unknown |
0.920 |
-0.137 |
| 39 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.912 |
0.069 |
| 40 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.906 |
-0.099 |
| 41 |
248505_at |
AT5G50360
|
unknown |
0.906 |
-0.007 |
| 42 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.902 |
-0.335 |
| 43 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.900 |
0.047 |
| 44 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.900 |
0.178 |
| 45 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.898 |
-0.084 |
| 46 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.890 |
0.146 |
| 47 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.889 |
0.126 |
| 48 |
249246_at |
AT5G42290
|
[transcription activator-related] |
0.879 |
-0.212 |
| 49 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
0.877 |
0.052 |
| 50 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.867 |
0.137 |
| 51 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.866 |
-0.036 |
| 52 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.852 |
0.165 |
| 53 |
256789_at |
AT3G13672
|
SINA2 |
0.852 |
0.079 |
| 54 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
0.850 |
0.359 |
| 55 |
264494_at |
AT1G27461
|
unknown |
0.842 |
0.206 |
| 56 |
250826_at |
AT5G05220
|
unknown |
0.832 |
0.003 |
| 57 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.829 |
0.157 |
| 58 |
258878_at |
AT3G03170
|
unknown |
0.815 |
-0.019 |
| 59 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.803 |
-0.055 |
| 60 |
245007_at |
ATCG00350
|
PSAA |
0.799 |
-0.024 |
| 61 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.797 |
0.075 |
| 62 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.790 |
-0.385 |
| 63 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.788 |
-0.057 |
| 64 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.787 |
0.079 |
| 65 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.785 |
0.055 |
| 66 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.780 |
0.037 |
| 67 |
250624_at |
AT5G07330
|
unknown |
0.779 |
-0.189 |
| 68 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
0.773 |
-0.077 |
| 69 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.768 |
-0.070 |
| 70 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.767 |
0.084 |
| 71 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.756 |
0.075 |
| 72 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.755 |
-0.153 |
| 73 |
259015_at |
AT3G07350
|
unknown |
0.748 |
0.131 |
| 74 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.741 |
-0.273 |
| 75 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.741 |
0.237 |
| 76 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.717 |
0.291 |
| 77 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.717 |
0.043 |
| 78 |
257670_at |
AT3G20340
|
unknown |
0.714 |
0.161 |
| 79 |
252073_at |
AT3G51750
|
unknown |
0.712 |
-0.117 |
| 80 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.706 |
0.128 |
| 81 |
254823_at |
AT4G12580
|
unknown |
0.699 |
0.094 |
| 82 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.699 |
0.051 |
| 83 |
252321_at |
AT3G48510
|
unknown |
0.697 |
-0.261 |
| 84 |
266990_at |
AT2G39190
|
ATATH8 |
0.691 |
0.136 |
| 85 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.690 |
0.198 |
| 86 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
0.689 |
-0.037 |
| 87 |
254178_at |
AT4G23880
|
unknown |
0.689 |
0.107 |
| 88 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.683 |
-0.040 |
| 89 |
257085_at |
AT3G20630
|
ATUBP14, PER1, phosphate deficiency root hair defective1, TTN6, TITAN6, UBP14, ubiquitin-specific protease 14 |
0.680 |
0.054 |
| 90 |
248642_at |
AT5G49120
|
unknown |
0.680 |
0.251 |
| 91 |
246299_at |
AT3G51810
|
AT3, ATEM1, ARABIDOPSIS THALIANA LATE EMBRYOGENESIS ABUNDANT 1, EM1, LATE EMBRYOGENESIS ABUNDANT 1, GEA1, GUANINE NUCLEOTIDE EXCHANGE FACTOR FOR ADP RIBOSYLATION FACTORS 1 |
0.677 |
0.111 |
| 92 |
264518_at |
AT1G09980
|
[Putative serine esterase family protein] |
0.676 |
-0.175 |
| 93 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.675 |
0.063 |
| 94 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.673 |
0.054 |
| 95 |
257859_at |
AT3G12955
|
SAUR74, SMALL AUXIN UPREGULATED RNA 74 |
0.670 |
0.356 |
| 96 |
252100_at |
AT3G51110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.667 |
0.186 |
| 97 |
267429_at |
AT2G34850
|
MEE25, maternal effect embryo arrest 25 |
0.661 |
0.100 |
| 98 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.657 |
0.191 |
| 99 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
0.655 |
0.029 |
| 100 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
0.653 |
0.110 |
| 101 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.651 |
-0.217 |
| 102 |
255049_at |
AT4G09610
|
GASA2, GAST1 protein homolog 2 |
0.642 |
0.045 |
| 103 |
255943_at |
AT1G22370
|
AtUGT85A5, UDP-glucosyl transferase 85A5, UGT85A5, UDP-glucosyl transferase 85A5 |
0.642 |
0.159 |
| 104 |
262049_at |
AT1G80180
|
unknown |
0.637 |
0.179 |
| 105 |
267511_at |
AT2G45670
|
[calcineurin B subunit-related] |
0.634 |
0.002 |
| 106 |
248276_at |
AT5G53550
|
ATYSL3, YELLOW STRIPE LIKE 3, YSL3, YELLOW STRIPE like 3 |
0.631 |
0.204 |
| 107 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
0.630 |
0.050 |
| 108 |
248218_at |
AT5G53710
|
unknown |
0.630 |
0.141 |
| 109 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
0.628 |
0.034 |
| 110 |
254597_at |
AT4G18980
|
AtS40-3, AtS40-3 |
0.626 |
0.498 |
| 111 |
267538_at |
AT2G41870
|
[Remorin family protein] |
0.621 |
0.157 |
| 112 |
263934_at |
AT2G35950
|
EDA12, embryo sac development arrest 12 |
0.620 |
0.056 |
| 113 |
260010_at |
AT1G68020
|
ATTPS6, TPS6, TREHALOSE -6-PHOSPHATASE SYNTHASE S6 |
0.619 |
-0.028 |
| 114 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.615 |
-0.109 |
| 115 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.614 |
0.062 |
| 116 |
259314_at |
AT3G05260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.614 |
0.142 |
| 117 |
263931_at |
AT2G36220
|
unknown |
0.612 |
-0.114 |
| 118 |
250937_at |
AT5G03230
|
unknown |
0.612 |
-0.163 |
| 119 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
0.608 |
0.553 |
| 120 |
261567_at |
AT1G33055
|
unknown |
0.606 |
0.007 |
| 121 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.602 |
-0.020 |
| 122 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.596 |
-0.102 |
| 123 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.594 |
-0.206 |
| 124 |
259426_at |
AT1G01470
|
LEA14, LATE EMBRYOGENESIS ABUNDANT 14, LSR3, LIGHT STRESS-REGULATED 3 |
0.588 |
0.004 |
| 125 |
253881_at |
AT4G27640
|
[ARM repeat superfamily protein] |
0.588 |
0.021 |
| 126 |
255602_at |
AT4G01026
|
PYL7, PYR1-like 7, RCAR2, regulatory components of ABA receptor 2 |
0.579 |
0.025 |
| 127 |
245831_at |
AT1G48840
|
unknown |
0.579 |
-0.019 |
| 128 |
245897_at |
AT5G09400
|
KUP7, K+ uptake permease 7 |
0.576 |
-0.048 |
| 129 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
0.568 |
0.040 |
| 130 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.567 |
0.281 |
| 131 |
262803_at |
AT1G21000
|
[PLATZ transcription factor family protein] |
0.565 |
0.030 |
| 132 |
266019_at |
AT2G18750
|
[Calmodulin-binding protein] |
0.559 |
0.129 |
| 133 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.558 |
-0.100 |
| 134 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
0.554 |
-0.159 |
| 135 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
0.552 |
-0.082 |
| 136 |
263605_at |
AT2G16485
|
NERD, Needed for RDR2-independent DNA methylation |
0.550 |
0.082 |
| 137 |
249944_at |
AT5G22290
|
anac089, NAC domain containing protein 89, FSQ6, fructose-sensing quantitative trait locus 6, NAC089, NAC domain containing protein 89 |
0.545 |
0.053 |
| 138 |
258026_at |
AT3G19290
|
ABF4, ABRE binding factor 4, AREB2, ABA-RESPONSIVE ELEMENT BINDING PROTEIN 2 |
0.544 |
-0.017 |
| 139 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.544 |
0.216 |
| 140 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
0.543 |
0.010 |
| 141 |
261863_at |
AT1G50630
|
unknown |
0.542 |
-0.004 |
| 142 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.542 |
0.020 |
| 143 |
247511_at |
AT5G62040
|
BFT, brother of FT and TFL1 |
0.538 |
-0.223 |
| 144 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
0.538 |
-0.253 |
| 145 |
247585_at |
AT5G60680
|
unknown |
0.538 |
-0.013 |
| 146 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.537 |
0.015 |
| 147 |
253859_at |
AT4G27657
|
unknown |
0.537 |
-0.041 |
| 148 |
262452_at |
AT1G11210
|
unknown |
0.529 |
0.059 |
| 149 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.528 |
-0.037 |
| 150 |
251829_at |
AT3G55020
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.522 |
-0.265 |
| 151 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.521 |
0.071 |
| 152 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.520 |
-0.018 |
| 153 |
262322_at |
AT1G27590
|
unknown |
0.519 |
0.038 |
| 154 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.518 |
-0.146 |
| 155 |
246215_at |
AT4G37180
|
[Homeodomain-like superfamily protein] |
0.518 |
0.066 |
| 156 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.517 |
0.061 |
| 157 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
0.515 |
0.095 |
| 158 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
0.514 |
0.572 |
| 159 |
247421_at |
AT5G62800
|
[Protein with RING/U-box and TRAF-like domains] |
0.511 |
0.208 |
| 160 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.509 |
0.022 |
| 161 |
253322_at |
AT4G33980
|
unknown |
0.508 |
0.071 |
| 162 |
249937_at |
AT5G22470
|
[NAD+ ADP-ribosyltransferases] |
0.508 |
0.096 |
| 163 |
262667_at |
AT1G62810
|
CuAO1, COPPER AMINE OXIDASE1 |
0.506 |
-0.031 |
| 164 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.504 |
0.171 |
| 165 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.503 |
0.007 |
| 166 |
265481_at |
AT2G15960
|
unknown |
0.503 |
0.061 |
| 167 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
0.502 |
0.178 |
| 168 |
259231_at |
AT3G11410
|
AHG3, ATPP2CA, ARABIDOPSIS THALIANA PROTEIN PHOSPHATASE 2CA, PP2CA, protein phosphatase 2CA |
0.502 |
0.049 |
| 169 |
247490_at |
AT5G61850
|
LFY, LEAFY, LFY3, LEAFY 3 |
0.501 |
0.061 |
| 170 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.501 |
0.328 |
| 171 |
261947_at |
AT1G64470
|
[Ubiquitin-like superfamily protein] |
0.500 |
0.001 |
| 172 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
0.500 |
-0.076 |
| 173 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
0.498 |
0.031 |
| 174 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.497 |
0.040 |
| 175 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.491 |
0.203 |
| 176 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.490 |
0.064 |
| 177 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.484 |
-0.087 |
| 178 |
254169_at |
AT4G24290
|
[MAC/Perforin domain-containing protein] |
0.483 |
0.099 |
| 179 |
263986_at |
AT2G42790
|
CSY3, citrate synthase 3 |
0.483 |
0.015 |
| 180 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
0.482 |
-0.065 |
| 181 |
250935_at |
AT5G03240
|
UBQ3, polyubiquitin 3 |
0.481 |
0.015 |
| 182 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.480 |
0.032 |
| 183 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.480 |
0.049 |
| 184 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.478 |
-0.156 |
| 185 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.477 |
0.088 |
| 186 |
263751_at |
AT2G21300
|
[ATP binding microtubule motor family protein] |
0.475 |
-0.040 |
| 187 |
261496_at |
AT1G28360
|
ATERF12, ERF DOMAIN PROTEIN 12, ERF12, ERF domain protein 12 |
0.474 |
0.045 |
| 188 |
264480_at |
AT1G77300
|
ASHH2, ASH1 HOMOLOG 2, CCR1, CAROTENOID CHLOROPLAST REGULATORY1, EFS, EARLY FLOWERING IN SHORT DAYS, LAZ2, LAZARUS 2, SDG8, SET DOMAIN GROUP 8 |
0.473 |
0.113 |
| 189 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.471 |
-0.037 |
| 190 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
0.471 |
0.004 |
| 191 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.468 |
-0.005 |
| 192 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
0.466 |
0.020 |
| 193 |
259012_at |
AT3G07360
|
ATPUB9, ARABIDOPSIS THALIANA PLANT U-BOX 9, PUB9, plant U-box 9 |
0.463 |
-0.165 |
| 194 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.463 |
-0.171 |
| 195 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.460 |
0.239 |
| 196 |
250938_at |
AT5G03180
|
[RING/U-box superfamily protein] |
0.458 |
-0.567 |
| 197 |
264167_at |
AT1G02060
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.458 |
0.012 |
| 198 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.457 |
0.299 |
| 199 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.456 |
-0.171 |
| 200 |
258652_at |
AT3G09910
|
ATRAB18C, ATRABC2B, RAB GTPase homolog C2B, RABC2b, RAB GTPase homolog C2B |
0.451 |
-0.282 |
| 201 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.450 |
-0.148 |
| 202 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.449 |
0.446 |
| 203 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
0.448 |
0.190 |
| 204 |
250809_at |
AT5G05140
|
[Transcription elongation factor (TFIIS) family protein] |
0.448 |
0.105 |
| 205 |
260875_at |
AT1G21410
|
SKP2A |
0.447 |
0.115 |
| 206 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.447 |
0.026 |
| 207 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
0.447 |
-0.167 |
| 208 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
0.447 |
0.063 |
| 209 |
250858_at |
AT5G04760
|
[Duplicated homeodomain-like superfamily protein] |
0.443 |
0.132 |
| 210 |
252057_at |
AT3G52480
|
unknown |
0.441 |
0.134 |
| 211 |
258374_at |
AT3G14360
|
[alpha/beta-Hydrolases superfamily protein] |
0.438 |
-0.278 |
| 212 |
258382_at |
AT3G15355
|
PFU1, PHO2 FAMILY UBIQUITIN CONJUGATION ENZYME 1, UBC25, ubiquitin-conjugating enzyme 25 |
0.438 |
0.006 |
| 213 |
253672_at |
AT4G29820
|
ATCFIM-25, ARABIDOPSIS THALIANA HOMOLOG OF CFIM-25, CFIM-25, homolog of CFIM-25 |
0.438 |
-0.281 |
| 214 |
264314_at |
AT1G70420
|
unknown |
0.437 |
0.159 |
| 215 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.437 |
0.192 |
| 216 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.436 |
0.179 |
| 217 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.436 |
0.145 |
| 218 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.435 |
-0.105 |
| 219 |
245070_at |
AT2G23240
|
AtMT4b, Arabidopsis thaliana metallothionein 4b |
0.434 |
0.141 |
| 220 |
255640_at |
AT4G00800
|
SETH5 |
0.434 |
-0.162 |
| 221 |
244967_at |
ATCG00630
|
PSAJ |
0.433 |
0.099 |
| 222 |
251156_at |
AT3G63060
|
EDL3, EID1-like 3 |
0.433 |
0.201 |
| 223 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
0.433 |
0.009 |
| 224 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.428 |
-0.253 |
| 225 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
0.428 |
0.101 |
| 226 |
249134_at |
AT5G43150
|
unknown |
0.427 |
-0.450 |
| 227 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.427 |
-0.214 |
| 228 |
257709_at |
AT3G27325
|
[hydrolases, acting on ester bonds] |
0.427 |
0.053 |
| 229 |
265276_at |
AT2G28400
|
unknown |
0.425 |
0.266 |
| 230 |
247374_at |
AT5G63190
|
[MA3 domain-containing protein] |
0.424 |
-0.008 |
| 231 |
258939_at |
AT3G10020
|
unknown |
0.424 |
-0.070 |
| 232 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
0.424 |
0.133 |
| 233 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
0.424 |
-0.350 |
| 234 |
264836_at |
AT1G03610
|
unknown |
0.421 |
-0.018 |
| 235 |
249905_at |
AT5G22720
|
[F-box/RNI-like superfamily protein] |
0.420 |
-0.044 |
| 236 |
261711_at |
AT1G32700
|
[PLATZ transcription factor family protein] |
0.420 |
0.057 |
| 237 |
262061_at |
AT1G80110
|
ATPP2-B11, phloem protein 2-B11, PP2-B11, phloem protein 2-B11 |
0.419 |
0.063 |
| 238 |
248558_at |
AT5G49990
|
[Xanthine/uracil permease family protein] |
0.419 |
-0.418 |
| 239 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
0.418 |
0.139 |
| 240 |
251580_at |
AT3G58450
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.416 |
0.189 |
| 241 |
251132_at |
AT5G01200
|
[Duplicated homeodomain-like superfamily protein] |
0.416 |
-0.284 |
| 242 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
0.416 |
-0.259 |
| 243 |
259004_at |
AT3G01840
|
LYK2, LysM-containing receptor-like kinase 2 |
0.416 |
0.524 |
| 244 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
0.414 |
0.147 |
| 245 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.414 |
0.327 |
| 246 |
265681_at |
AT2G24370
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.413 |
0.136 |
| 247 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
0.413 |
0.187 |
| 248 |
260345_at |
AT1G69270
|
AtRPK1, RPK1, receptor-like protein kinase 1 |
0.411 |
0.107 |
| 249 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
0.410 |
0.668 |
| 250 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.410 |
0.092 |
| 251 |
263907_at |
AT2G36270
|
ABI5, ABA INSENSITIVE 5, AtABI5, GIA1, GROWTH-INSENSITIVITY TO ABA 1 |
0.407 |
0.009 |
| 252 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.407 |
0.096 |
| 253 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.406 |
0.075 |
| 254 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.406 |
-0.032 |
| 255 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.403 |
0.253 |
| 256 |
262788_at |
AT1G10690
|
unknown |
0.403 |
0.217 |
| 257 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.403 |
-0.095 |
| 258 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.402 |
0.052 |
| 259 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.397 |
0.122 |
| 260 |
246782_at |
AT5G27320
|
ATGID1C, GID1C, GA INSENSITIVE DWARF1C |
0.397 |
0.019 |
| 261 |
245477_at |
AT4G16110
|
ARR2, response regulator 2, RR2, response regulator 2 |
0.394 |
-0.044 |
| 262 |
260878_at |
AT1G21450
|
SCL1, SCARECROW-like 1 |
0.393 |
0.123 |
| 263 |
246012_at |
AT5G10650
|
[RING/U-box superfamily protein] |
0.393 |
0.137 |
| 264 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.392 |
0.099 |
| 265 |
263545_at |
AT2G21560
|
unknown |
0.392 |
0.355 |
| 266 |
245412_at |
AT4G17280
|
[Auxin-responsive family protein] |
0.391 |
0.198 |
| 267 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
0.391 |
-0.001 |
| 268 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.390 |
0.100 |
| 269 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.390 |
-0.103 |
| 270 |
259500_at |
AT1G15740
|
[Leucine-rich repeat family protein] |
0.390 |
0.102 |
| 271 |
260627_at |
AT1G62310
|
[transcription factor jumonji (jmjC) domain-containing protein] |
0.388 |
0.211 |
| 272 |
250881_at |
AT5G04080
|
unknown |
0.388 |
-0.004 |
| 273 |
264957_at |
AT1G77000
|
ATSKP2;2, ARABIDOPSIS HOMOLOG OF HOMOLOG OF HUMAN SKP2 2, SKP2B |
0.387 |
0.220 |
| 274 |
259856_at |
AT1G68440
|
unknown |
0.387 |
0.082 |
| 275 |
257224_at |
AT3G27870
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.387 |
-0.049 |
| 276 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.384 |
0.304 |
| 277 |
264889_at |
AT1G23050
|
hydroxyproline-rich glycoprotein family protein |
0.383 |
0.164 |
| 278 |
246889_at |
AT5G25470
|
[AP2/B3-like transcriptional factor family protein] |
0.383 |
0.020 |
| 279 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.383 |
0.040 |
| 280 |
264779_at |
AT1G08570
|
ACHT4, atypical CYS HIS rich thioredoxin 4 |
0.382 |
0.062 |
| 281 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
0.381 |
-0.238 |
| 282 |
262436_at |
AT1G47610
|
[Transducin/WD40 repeat-like superfamily protein] |
0.381 |
0.038 |
| 283 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.381 |
0.243 |
| 284 |
247992_at |
AT5G56520
|
unknown |
0.381 |
-0.086 |
| 285 |
265767_at |
AT2G48110
|
MED33B, REF4, REDUCED EPIDERMAL FLUORESCENCE 4 |
0.380 |
0.038 |
| 286 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.379 |
0.101 |
| 287 |
265731_at |
AT2G01130
|
[DEA(D/H)-box RNA helicase family protein] |
0.379 |
-0.106 |
| 288 |
262199_at |
AT1G53800
|
unknown |
0.379 |
0.009 |
| 289 |
258874_at |
AT3G03230
|
[alpha/beta-Hydrolases superfamily protein] |
0.379 |
0.271 |
| 290 |
247004_at |
AT5G67570
|
DG1, DELAYED GREENING 1, EMB1408, embryo defective 1408, EMB246, EMBRYO DEFECTIVE 246 |
0.377 |
0.184 |
| 291 |
261446_at |
AT1G21170
|
SEC5B |
0.376 |
0.102 |
| 292 |
255673_at |
AT4G00340
|
RLK4, receptor-like protein kinase 4 |
0.376 |
0.122 |
| 293 |
257122_at |
AT3G20250
|
APUM5, pumilio 5, PUM5, pumilio 5 |
0.374 |
0.004 |
| 294 |
263265_at |
AT2G38820
|
unknown |
0.373 |
0.216 |
| 295 |
252179_at |
AT3G50760
|
GATL2, galacturonosyltransferase-like 2 |
0.373 |
-0.034 |
| 296 |
250737_at |
AT5G06370
|
[NC domain-containing protein-related] |
0.371 |
-0.015 |
| 297 |
251859_at |
AT3G54680
|
[proteophosphoglycan-related] |
0.368 |
0.201 |
| 298 |
247863_at |
AT5G57900
|
SKIP1, SKP1 interacting partner 1 |
0.368 |
0.070 |
| 299 |
252938_at |
AT4G39190
|
unknown |
0.367 |
0.156 |
| 300 |
266583_at |
AT2G46220
|
[Uncharacterized conserved protein (DUF2358)] |
0.366 |
0.039 |
| 301 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
-0.849 |
-0.005 |
| 302 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.805 |
0.080 |
| 303 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.777 |
0.164 |
| 304 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.776 |
0.029 |
| 305 |
248028_at |
AT5G55620
|
unknown |
-0.748 |
0.170 |
| 306 |
261979_at |
AT1G37130
|
ATNR2, ARABIDOPSIS NITRATE REDUCTASE 2, B29, CHL3, CHLORATE RESISTANT 3, NIA2, nitrate reductase 2, NIA2-1, NR, NITRATE REDUCTASE, NR2, NITRATE REDUCTASE 2 |
-0.748 |
0.267 |
| 307 |
258370_at |
AT3G14395
|
unknown |
-0.737 |
-0.234 |
| 308 |
244974_at |
ATCG00700
|
PSBN, photosystem II reaction center protein N |
-0.723 |
0.124 |
| 309 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.692 |
-0.029 |
| 310 |
253532_at |
AT4G31570
|
unknown |
-0.687 |
0.102 |
| 311 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.668 |
0.145 |
| 312 |
244934_at |
ATCG01080
|
NDHG |
-0.640 |
0.019 |
| 313 |
267545_at |
AT2G32690
|
ATGRP23, GLYCINE-RICH PROTEIN 23, GRP23, glycine-rich protein 23 |
-0.632 |
0.209 |
| 314 |
253800_at |
AT4G28160
|
[hydroxyproline-rich glycoprotein family protein] |
-0.613 |
-0.094 |
| 315 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.607 |
-0.004 |
| 316 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.603 |
-0.050 |
| 317 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.601 |
0.039 |
| 318 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.593 |
0.084 |
| 319 |
248350_at |
AT5G52160
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.588 |
0.211 |
| 320 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.587 |
0.132 |
| 321 |
245524_at |
AT4G15920
|
AtSWEET17, SWEET17 |
-0.575 |
0.009 |
| 322 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.574 |
0.257 |
| 323 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.567 |
-0.061 |
| 324 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.559 |
-0.088 |
| 325 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
-0.551 |
0.105 |
| 326 |
245008_at |
ATCG00360
|
YCF3 |
-0.545 |
0.036 |
| 327 |
262508_at |
AT1G11300
|
[protein serine/threonine kinases] |
-0.536 |
0.617 |
| 328 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.536 |
0.114 |
| 329 |
244903_at |
ATMG00660
|
ORF149 |
-0.536 |
0.117 |
| 330 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
-0.531 |
-0.048 |
| 331 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.530 |
-0.116 |
| 332 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.528 |
0.112 |
| 333 |
246498_at |
AT5G16230
|
[Plant stearoyl-acyl-carrier-protein desaturase family protein] |
-0.523 |
-0.053 |
| 334 |
257570_at |
AT3G13662
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.517 |
0.103 |
| 335 |
258289_at |
AT3G23450
|
unknown |
-0.507 |
0.114 |
| 336 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.506 |
0.069 |
| 337 |
264277_at |
AT1G60390
|
PG1, polygalacturonase 1 |
-0.497 |
0.057 |
| 338 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.485 |
-0.044 |
| 339 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.483 |
0.244 |
| 340 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.482 |
0.346 |
| 341 |
246419_at |
AT5G17030
|
UGT78D3, UDP-glucosyl transferase 78D3 |
-0.476 |
0.113 |
| 342 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
-0.470 |
0.167 |
| 343 |
249718_at |
AT5G35740
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.465 |
0.173 |
| 344 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.465 |
0.102 |
| 345 |
266325_at |
AT2G46630
|
unknown |
-0.457 |
0.050 |
| 346 |
252876_at |
AT4G39970
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.455 |
0.049 |
| 347 |
267636_at |
AT2G42110
|
unknown |
-0.453 |
0.280 |
| 348 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.449 |
0.081 |
| 349 |
263485_at |
AT2G29890
|
ATVLN1, VLN1, villin 1 |
-0.447 |
0.208 |
| 350 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.445 |
0.129 |
| 351 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.443 |
0.176 |
| 352 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.441 |
0.108 |
| 353 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.440 |
-0.321 |
| 354 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.439 |
0.075 |
| 355 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
-0.439 |
0.031 |
| 356 |
266549_at |
AT2G35150
|
EXL7, EXORDIUM LIKE 7 |
-0.438 |
0.266 |
| 357 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.438 |
-0.115 |
| 358 |
249515_at |
AT5G38530
|
TSBtype2, tryptophan synthase beta type 2 |
-0.436 |
-0.288 |
| 359 |
250269_at |
AT5G12970
|
[Calcium-dependent lipid-binding (CaLB domain) plant phosphoribosyltransferase family protein] |
-0.432 |
0.179 |
| 360 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.430 |
-0.088 |
| 361 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.428 |
0.156 |
| 362 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.427 |
-0.017 |
| 363 |
247977_at |
AT5G56850
|
unknown |
-0.426 |
0.160 |
| 364 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.422 |
0.136 |
| 365 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
-0.422 |
0.037 |
| 366 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
-0.422 |
0.058 |
| 367 |
248054_at |
AT5G55820
|
WYR, WYRD |
-0.416 |
-0.031 |
| 368 |
257697_at |
AT3G12700
|
NANA, NANA |
-0.413 |
0.311 |
| 369 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.413 |
0.006 |
| 370 |
245000_at |
ATCG00210
|
YCF6 |
-0.410 |
-0.010 |
| 371 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.408 |
0.118 |
| 372 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
-0.407 |
0.239 |
| 373 |
246475_at |
AT5G16720
|
MyoB3, myosin binding protein 3 |
-0.405 |
0.141 |
| 374 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.404 |
0.052 |
| 375 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.403 |
0.143 |
| 376 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
-0.402 |
0.213 |
| 377 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
-0.401 |
-0.040 |
| 378 |
265873_at |
AT2G01630
|
[O-Glycosyl hydrolases family 17 protein] |
-0.398 |
-0.126 |
| 379 |
264801_at |
AT1G08840
|
emb2411, embryo defective 2411 |
-0.397 |
0.061 |
| 380 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
-0.396 |
0.101 |
| 381 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.394 |
0.191 |
| 382 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.393 |
0.070 |
| 383 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.390 |
0.036 |
| 384 |
257825_at |
AT3G26700
|
[Protein kinase superfamily protein] |
-0.390 |
-0.048 |
| 385 |
260975_at |
AT1G53430
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.388 |
0.272 |
| 386 |
244933_at |
ATCG01070
|
NDHE |
-0.387 |
0.039 |
| 387 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.384 |
0.049 |
| 388 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
-0.383 |
0.203 |
| 389 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
-0.383 |
-0.053 |
| 390 |
262558_at |
AT1G31335
|
unknown |
-0.383 |
0.104 |
| 391 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.382 |
0.159 |
| 392 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.382 |
-0.158 |
| 393 |
255790_at |
AT2G33560
|
BUBR1, BUB1-related (BUB1: budding uninhibited by benzymidazol 1) |
-0.380 |
0.106 |
| 394 |
244972_at |
ATCG00680
|
PSBB, photosystem II reaction center protein B |
-0.380 |
0.063 |
| 395 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.379 |
0.033 |
| 396 |
255955_at |
AT1G22030
|
unknown |
-0.379 |
0.154 |
| 397 |
246200_at |
AT4G37240
|
unknown |
-0.376 |
0.247 |
| 398 |
267192_at |
AT2G30890
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.374 |
0.206 |
| 399 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-0.372 |
-0.009 |
| 400 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.371 |
0.011 |
| 401 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.370 |
0.058 |
| 402 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.367 |
-0.012 |
| 403 |
259000_at |
AT3G01860
|
unknown |
-0.367 |
0.093 |
| 404 |
256163_at |
AT1G48820
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.367 |
0.135 |
| 405 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.367 |
0.071 |
| 406 |
249029_at |
AT5G44870
|
LAZ5, LAZARUS 5, TTR1, tolerance to Tobacco ringspot virus 1 |
-0.364 |
0.004 |
| 407 |
248835_at |
AT5G47250
|
[LRR and NB-ARC domains-containing disease resistance protein] |
-0.364 |
-0.005 |
| 408 |
263828_at |
AT2G40250
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.363 |
0.009 |
| 409 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
-0.361 |
0.169 |
| 410 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
-0.360 |
0.177 |
| 411 |
250482_at |
AT5G10320
|
unknown |
-0.359 |
-0.035 |
| 412 |
248582_at |
AT5G49910
|
cpHsc70-2, chloroplast heat shock protein 70-2, HSC70-7, HEAT SHOCK PROTEIN 70-7 |
-0.359 |
0.004 |
| 413 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.358 |
0.310 |
| 414 |
253978_at |
AT4G26660
|
unknown |
-0.358 |
0.087 |
| 415 |
264624_at |
AT1G08930
|
ERD6, EARLY RESPONSE TO DEHYDRATION 6 |
-0.358 |
0.085 |
| 416 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.357 |
0.052 |
| 417 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.355 |
-0.057 |
| 418 |
254145_at |
AT4G24700
|
unknown |
-0.355 |
-0.052 |
| 419 |
247567_at |
AT5G61190
|
[putative endonuclease or glycosyl hydrolase with C2H2-type zinc finger domain] |
-0.353 |
0.053 |
| 420 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-0.353 |
0.259 |
| 421 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
-0.353 |
0.013 |
| 422 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.353 |
0.218 |
| 423 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
-0.352 |
-0.129 |
| 424 |
251900_at |
AT3G54430
|
SRS6, SHI-related sequence 6 |
-0.352 |
0.228 |
| 425 |
254303_at |
AT4G22830
|
unknown |
-0.351 |
0.194 |
| 426 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.351 |
0.082 |
| 427 |
256518_at |
AT1G66080
|
unknown |
-0.351 |
-0.095 |
| 428 |
262873_at |
AT1G64700
|
unknown |
-0.349 |
0.108 |
| 429 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.349 |
0.236 |
| 430 |
254208_at |
AT4G24175
|
unknown |
-0.349 |
0.104 |
| 431 |
248865_at |
AT5G46790
|
PYL1, PYR1-like 1, RCAR12, regulatory components of ABA receptor 12 |
-0.348 |
0.075 |
| 432 |
263470_at |
AT2G31900
|
ATMYO5, MYOSIN 5, ATXIF, MYOSIN XI F, XIF, myosin-like protein XIF |
-0.348 |
0.028 |
| 433 |
255716_at |
AT4G00330
|
CRCK2, calmodulin-binding receptor-like cytoplasmic kinase 2 |
-0.347 |
0.148 |
| 434 |
245607_at |
AT4G14330
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.346 |
-0.125 |
| 435 |
262286_at |
AT1G68585
|
unknown |
-0.346 |
-0.078 |
| 436 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.346 |
0.203 |
| 437 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.345 |
0.071 |
| 438 |
259815_at |
AT1G49870
|
unknown |
-0.344 |
0.017 |
| 439 |
246386_at |
AT1G77410
|
BGAL16, beta-galactosidase 16 |
-0.344 |
0.046 |
| 440 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.344 |
-0.206 |
| 441 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.344 |
0.150 |
| 442 |
259250_at |
AT3G07580
|
unknown |
-0.344 |
-0.030 |
| 443 |
261175_at |
AT1G04800
|
[glycine-rich protein] |
-0.340 |
0.073 |
| 444 |
259912_at |
AT1G72670
|
iqd8, IQ-domain 8 |
-0.340 |
0.095 |
| 445 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.338 |
0.084 |
| 446 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
-0.336 |
0.070 |
| 447 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
-0.335 |
0.305 |
| 448 |
265376_at |
AT2G05810
|
[ARM repeat superfamily protein] |
-0.335 |
-0.030 |
| 449 |
262681_at |
AT1G75890
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.334 |
0.109 |
| 450 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.334 |
0.188 |
| 451 |
247959_at |
AT5G57080
|
unknown |
-0.333 |
-0.049 |
| 452 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.331 |
0.072 |
| 453 |
261384_at |
AT1G05440
|
[C-8 sterol isomerases] |
-0.329 |
0.153 |
| 454 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.328 |
0.498 |
| 455 |
246321_at |
AT1G16640
|
[AP2/B3-like transcriptional factor family protein] |
-0.327 |
0.118 |
| 456 |
251966_at |
AT3G53380
|
LecRK-VIII.1, L-type lectin receptor kinase VIII.1 |
-0.327 |
0.089 |
| 457 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.327 |
0.081 |
| 458 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-0.326 |
0.143 |
| 459 |
259363_at |
AT1G13270
|
MAP1B, METHIONINE AMINOPEPTIDASE 1B, MAP1C, methionine aminopeptidase 1B |
-0.326 |
-0.148 |
| 460 |
260685_at |
AT1G17650
|
AtGLYR2, GLYR2, glyoxylate reductase 2, GR2, GLYOXYLATE REDUCTASE 2 |
-0.324 |
0.165 |
| 461 |
250284_at |
AT5G13290
|
CRN, CORYNE, SOL2, SUPPRESSOR OF LLP1 2 |
-0.322 |
-0.186 |
| 462 |
265646_at |
AT2G27360
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.322 |
0.010 |
| 463 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
-0.320 |
0.129 |
| 464 |
249355_at |
AT5G40500
|
unknown |
-0.319 |
-0.035 |
| 465 |
267367_at |
AT2G44210
|
unknown |
-0.319 |
-0.127 |
| 466 |
248691_at |
AT5G48310
|
unknown |
-0.319 |
0.097 |
| 467 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
-0.319 |
-0.007 |
| 468 |
247400_at |
AT5G62840
|
[Phosphoglycerate mutase family protein] |
-0.318 |
0.071 |
| 469 |
267013_at |
AT2G39180
|
ATCRR2, CCR2, CRINKLY4 related 2 |
-0.316 |
-0.030 |
| 470 |
264473_at |
AT1G67180
|
[zinc finger (C3HC4-type RING finger) family protein / BRCT domain-containing protein] |
-0.316 |
-0.031 |
| 471 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.316 |
0.166 |
| 472 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.315 |
0.111 |
| 473 |
255817_at |
AT2G33330
|
PDLP3, plasmodesmata-located protein 3 |
-0.314 |
-0.118 |
| 474 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
-0.312 |
0.212 |
| 475 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
-0.311 |
-0.030 |
| 476 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.310 |
0.012 |
| 477 |
267618_at |
AT2G26760
|
CYCB1;4, Cyclin B1;4 |
-0.310 |
0.102 |
| 478 |
259928_at |
AT1G34380
|
[5'-3' exonuclease family protein] |
-0.310 |
-0.174 |
| 479 |
265964_at |
AT2G37510
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.310 |
0.052 |
| 480 |
258039_at |
AT3G21200
|
GluTRBP, GluTR binding protein, PGR7, proton gradient regulation 7 |
-0.310 |
0.085 |
| 481 |
246137_at |
AT5G28490
|
LSH1, LIGHT-DEPENDENT SHORT HYPOCOTYLS 1, OBO2, ORGAN BOUNDARY 2 |
-0.309 |
0.123 |
| 482 |
250837_at |
AT5G04620
|
ATBIOF, biotin F, BIO4, biotin 4, BIOF, biotin F |
-0.308 |
-0.018 |
| 483 |
264873_at |
AT1G24100
|
UGT74B1, UDP-glucosyl transferase 74B1 |
-0.308 |
0.062 |
| 484 |
258464_at |
AT3G17360
|
POK1, phragmoplast orienting kinesin 1 |
-0.307 |
0.020 |
| 485 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.307 |
0.164 |
| 486 |
252712_at |
AT3G43800
|
ATGSTU27, glutathione S-transferase tau 27, GSTU27, glutathione S-transferase tau 27 |
-0.307 |
0.070 |
| 487 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
-0.307 |
-0.092 |
| 488 |
247486_at |
AT5G62140
|
unknown |
-0.307 |
0.096 |
| 489 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
-0.307 |
-0.137 |
| 490 |
262693_at |
AT1G62780
|
unknown |
-0.307 |
0.171 |
| 491 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.306 |
0.141 |
| 492 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.306 |
0.098 |
| 493 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.305 |
0.114 |
| 494 |
262277_at |
AT1G68650
|
[Uncharacterized protein family (UPF0016)] |
-0.305 |
-0.089 |
| 495 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
-0.305 |
0.152 |
| 496 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.304 |
0.226 |
| 497 |
255876_at |
AT2G40480
|
unknown |
-0.304 |
0.012 |
| 498 |
266733_at |
AT2G03280
|
[O-fucosyltransferase family protein] |
-0.301 |
0.034 |
| 499 |
260332_at |
AT1G70470
|
unknown |
-0.301 |
0.313 |
| 500 |
251718_at |
AT3G56100
|
IMK3, MRLK, meristematic receptor-like kinase |
-0.300 |
0.010 |
| 501 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.300 |
0.075 |
| 502 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
-0.299 |
0.158 |
| 503 |
254466_at |
AT4G20430
|
[Subtilase family protein] |
-0.298 |
0.088 |
| 504 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
-0.297 |
0.208 |
| 505 |
247091_at |
AT5G66390
|
PRX72, PEROXIDASE 72 |
-0.297 |
0.329 |
| 506 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.296 |
0.131 |
| 507 |
244995_at |
ATCG00150
|
ATPI |
-0.295 |
0.030 |
| 508 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
-0.295 |
0.073 |
| 509 |
252786_at |
AT3G42670
|
CHR38, chromatin remodeling 38, CLSY, CLASSY1, CLSY1, CLASSY 1 |
-0.295 |
0.104 |
| 510 |
259607_at |
AT1G27940
|
ABCB13, ATP-binding cassette B13, PGP13, P-glycoprotein 13 |
-0.294 |
0.074 |
| 511 |
247826_at |
AT5G58480
|
[O-Glycosyl hydrolases family 17 protein] |
-0.293 |
0.158 |
| 512 |
253757_at |
AT4G28950
|
ARAC7, Arabidopsis RAC-like 7, ATRAC7, ATROP9, RAC7, ROP9, RHO-related protein from plants 9 |
-0.292 |
-0.201 |
| 513 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
-0.292 |
-0.168 |
| 514 |
252478_at |
AT3G46540
|
[ENTH/VHS family protein] |
-0.291 |
0.087 |
| 515 |
262081_at |
AT1G59540
|
ZCF125 |
-0.291 |
0.038 |
| 516 |
263864_at |
AT2G04530
|
CPZ, TRZ2, TRNASE Z 2 |
-0.290 |
0.122 |
| 517 |
248167_at |
AT5G54530
|
unknown |
-0.290 |
0.094 |
| 518 |
262338_at |
AT1G64185
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.289 |
-0.028 |
| 519 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
-0.288 |
-0.114 |
| 520 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.286 |
0.105 |
| 521 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
-0.286 |
0.123 |
| 522 |
261235_x_at |
AT1G32840
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
-0.285 |
0.132 |
| 523 |
257517_at |
AT3G16330
|
unknown |
-0.285 |
-0.027 |
| 524 |
261011_at |
AT1G26340
|
ATCB5-A, ARABIDOPSIS CYTOCHROME B5 ISOFORM A, B5 #6, CB5-A, cytochrome B5 isoform A |
-0.285 |
0.035 |
| 525 |
250326_at |
AT5G12080
|
ATMSL10, MSL10, mechanosensitive channel of small conductance-like 10 |
-0.285 |
-0.072 |
| 526 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.284 |
0.162 |
| 527 |
266891_at |
AT2G44690
|
ARAC9, Arabidopsis RAC-like 9, ATROP8, RHO-RELATED PROTEIN FROM PLANTS 8, ROP8, RHO-RELATED PROTEIN FROM PLANTS 8 |
-0.284 |
0.136 |
| 528 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.284 |
0.083 |
| 529 |
256856_at |
AT3G15110
|
unknown |
-0.284 |
-0.034 |
| 530 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.283 |
0.123 |
| 531 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.283 |
0.059 |
| 532 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
-0.282 |
-0.014 |
| 533 |
249809_at |
AT5G23910
|
[ATP binding microtubule motor family protein] |
-0.282 |
-0.052 |
| 534 |
255432_at |
AT4G03330
|
ATSYP123, SYP123, syntaxin of plants 123 |
-0.282 |
-0.051 |
| 535 |
259764_at |
AT1G64280
|
ATNPR1, ARABIDOPSIS NONEXPRESSER OF PR GENES 1, NIM1, NON-INDUCIBLE IMMUNITY 1, NPR1, NONEXPRESSER OF PR GENES 1, SAI1, SALICYLIC ACID INSENSITIVE 1 |
-0.281 |
0.080 |
| 536 |
246411_at |
AT1G57770
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.281 |
0.006 |
| 537 |
250257_at |
AT5G13770
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.281 |
-0.077 |
| 538 |
266952_at |
AT2G34555
|
ATGA2OX3, gibberellin 2-oxidase 3, GA2OX3, gibberellin 2-oxidase 3 |
-0.280 |
0.046 |
| 539 |
245461_at |
AT4G17000
|
unknown |
-0.280 |
0.000 |
| 540 |
258951_at |
AT3G01380
|
[transferases] |
-0.280 |
-0.077 |
| 541 |
248061_at |
AT5G55340
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.280 |
0.168 |
| 542 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.279 |
0.065 |
| 543 |
257136_at |
AT3G12890
|
ASML2, activator of spomin::LUC2 |
-0.279 |
0.228 |
| 544 |
254112_at |
AT4G24970
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
-0.278 |
-0.603 |
| 545 |
250719_at |
AT5G06250
|
DPA4, DEVELOPMENT-RELATED PcG TARGET IN THE APEX 4 |
-0.278 |
0.060 |
| 546 |
252785_at |
AT3G42660
|
[transducin family protein / WD-40 repeat family protein] |
-0.276 |
0.044 |
| 547 |
260601_at |
AT1G55910
|
ZIP11, zinc transporter 11 precursor |
-0.276 |
-0.111 |
| 548 |
260395_at |
AT1G69780
|
ATHB13 |
-0.275 |
0.155 |
| 549 |
245291_at |
AT4G16155
|
[dihydrolipoyl dehydrogenases] |
-0.275 |
-0.201 |
| 550 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.274 |
-0.086 |
| 551 |
247304_at |
AT5G63850
|
AAP4, amino acid permease 4 |
-0.274 |
0.195 |
| 552 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.274 |
-0.082 |
| 553 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.274 |
0.122 |
| 554 |
251292_at |
AT3G61920
|
unknown |
-0.273 |
-0.014 |
| 555 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
-0.273 |
0.230 |
| 556 |
267012_at |
AT2G39220
|
PLA IIB, PLP6, PATATIN-like protein 6 |
-0.272 |
0.135 |
| 557 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.272 |
-0.109 |
| 558 |
267294_at |
AT2G23670
|
YCF37, homolog of Synechocystis YCF37 |
-0.272 |
0.102 |
| 559 |
251942_at |
AT3G53480
|
ABCG37, ATP-binding cassette G37, ATPDR9, PLEIOTROPIC DRUG RESISTANCE 9, PDR9, pleiotropic drug resistance 9, PIS1, polar auxin transport inhibitor sensitive 1 |
-0.272 |
-0.030 |
| 560 |
248658_at |
AT5G48600
|
ATCAP-C, ARABIDOPSIS THALIANA CHROMOSOME ASSOCIATED PROTEIN-C, ATSMC3, structural maintenance of chromosome 3, ATSMC4, ARABIDOPSIS THALIANA STRUCTURAL MAINTENANCE OF CHROMOSOME 4, SMC3, structural maintenance of chromosome 3 |
-0.271 |
0.049 |
| 561 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
-0.271 |
-0.079 |
| 562 |
247214_at |
AT5G64850
|
unknown |
-0.271 |
-0.096 |
| 563 |
248307_at |
AT5G52850
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.271 |
-0.043 |
| 564 |
266797_at |
AT2G22840
|
AtGRF1, growth-regulating factor 1, GRF1, growth-regulating factor 1 |
-0.270 |
0.005 |
| 565 |
259838_at |
AT1G52220
|
CURT1C, CURVATURE THYLAKOID 1C |
-0.269 |
0.087 |
| 566 |
248493_at |
AT5G51100
|
FSD2, Fe superoxide dismutase 2 |
-0.268 |
0.065 |
| 567 |
245701_at |
AT5G04140
|
FD-GOGAT, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE, GLS1, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1, GLU1, glutamate synthase 1, GLUS |
-0.268 |
0.395 |
| 568 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.268 |
0.071 |
| 569 |
252603_at |
AT3G45050
|
unknown |
-0.268 |
0.239 |
| 570 |
260455_at |
AT1G72500
|
[LOCATED IN: plasma membrane] |
-0.267 |
-0.059 |
| 571 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.267 |
0.239 |
| 572 |
258462_at |
AT3G17350
|
unknown |
-0.267 |
0.119 |
| 573 |
262878_at |
AT1G64770
|
NDF2, NDH-dependent cyclic electron flow 1, NDH45, NAD(P)H DEHYDROGENASE SUBUNIT 45, PnsB2, Photosynthetic NDH subcomplex B 2 |
-0.266 |
0.103 |
| 574 |
267219_at |
AT2G02590
|
unknown |
-0.266 |
0.107 |
| 575 |
246269_at |
AT4G37110
|
[Zinc-finger domain of monoamine-oxidase A repressor R1] |
-0.266 |
-0.010 |
| 576 |
250820_at |
AT5G05160
|
RUL1, REDUCED IN LATERAL GROWTH1 |
-0.265 |
0.081 |
| 577 |
261277_at |
AT1G20230
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.264 |
0.118 |
| 578 |
247689_at |
AT5G59770
|
[Protein-tyrosine phosphatase-like, PTPLA] |
-0.263 |
-0.230 |
| 579 |
265154_at |
AT1G30960
|
[GTP-binding family protein] |
-0.263 |
0.138 |
| 580 |
265844_at |
AT2G35620
|
FEI2, FEI 2 |
-0.262 |
-0.400 |
| 581 |
264397_at |
AT1G11820
|
[O-Glycosyl hydrolases family 17 protein] |
-0.262 |
-0.197 |
| 582 |
256400_at |
AT3G06140
|
LUL4, LOG2-LIKE UBIQUITIN LIGASE4 |
-0.262 |
0.025 |
| 583 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.262 |
-0.473 |
| 584 |
245389_at |
AT4G17480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.262 |
0.378 |
| 585 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
-0.261 |
0.209 |
| 586 |
262744_at |
AT1G28680
|
[HXXXD-type acyl-transferase family protein] |
-0.261 |
-0.349 |
| 587 |
257937_at |
AT3G19810
|
unknown |
-0.261 |
-0.168 |
| 588 |
245809_at |
AT1G58440
|
DRY2, DROUGHT HYPERSENSITIVE 2, SQE1, SQUALENE EPOXIDASE 1, XF1 |
-0.261 |
-0.109 |
| 589 |
248634_at |
AT5G49030
|
OVA2, ovule abortion 2 |
-0.261 |
-0.006 |
| 590 |
245690_at |
AT5G04230
|
ATPAL3, PAL3, phenyl alanine ammonia-lyase 3 |
-0.260 |
0.172 |
| 591 |
248854_at |
AT5G46580
|
[pentatricopeptide (PPR) repeat-containing protein] |
-0.260 |
-0.099 |
| 592 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
-0.259 |
0.061 |
| 593 |
259448_at |
AT1G13790
|
FDM4, factor of DNA methylation 4 |
-0.259 |
0.196 |
| 594 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.259 |
0.277 |
| 595 |
245867_at |
AT1G58080
|
ATATP-PRT1, ATP phosphoribosyl transferase 1, ATP-PRT1, ATP phosphoribosyl transferase 1, HISN1A |
-0.257 |
-0.222 |
| 596 |
245560_at |
AT4G15480
|
UGT84A1 |
-0.257 |
0.156 |
| 597 |
262486_at |
AT1G21740
|
unknown |
-0.257 |
0.117 |
| 598 |
264963_at |
AT1G60600
|
ABC4, ABERRANT CHLOROPLAST DEVELOPMENT 4 |
-0.257 |
0.186 |
| 599 |
258010_at |
AT3G19300
|
[Protein kinase superfamily protein] |
-0.256 |
-0.030 |
| 600 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.256 |
0.231 |