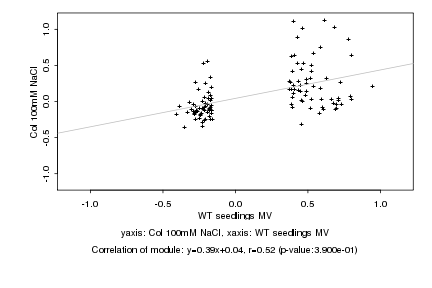
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT seedlings MV |
Log2 signal ratio
Col 100mM NaCl |
| 1 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.940 |
0.220 |
| 2 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.800 |
0.037 |
| 3 |
250826_at |
AT5G05220
|
unknown |
0.798 |
0.645 |
| 4 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.791 |
0.081 |
| 5 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.776 |
0.866 |
| 6 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.730 |
-0.033 |
| 7 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.724 |
0.276 |
| 8 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.709 |
0.027 |
| 9 |
256044_at |
AT1G07160
|
[Protein phosphatase 2C family protein] |
0.705 |
0.044 |
| 10 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.694 |
-0.095 |
| 11 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
0.693 |
-0.031 |
| 12 |
257918_at |
AT3G23230
|
AtERF98, AtTDR1, ERF98, Ethylene Response Factor 98, TDR1, Transcriptional Regulator of Defense Response 1 |
0.687 |
-0.097 |
| 13 |
253832_at |
AT4G27654
|
unknown |
0.679 |
1.030 |
| 14 |
251114_at |
AT5G01380
|
[Homeodomain-like superfamily protein] |
0.670 |
-0.014 |
| 15 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
0.658 |
0.032 |
| 16 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.628 |
0.323 |
| 17 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
0.612 |
1.136 |
| 18 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.606 |
-0.107 |
| 19 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.599 |
-0.081 |
| 20 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
0.588 |
0.034 |
| 21 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.586 |
0.190 |
| 22 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.585 |
0.753 |
| 23 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.578 |
-0.159 |
| 24 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.536 |
0.210 |
| 25 |
253859_at |
AT4G27657
|
unknown |
0.535 |
0.672 |
| 26 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
0.524 |
0.416 |
| 27 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.519 |
0.505 |
| 28 |
264314_at |
AT1G70420
|
unknown |
0.519 |
0.034 |
| 29 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.517 |
-0.095 |
| 30 |
255542_at |
AT4G01860
|
[Transducin family protein / WD-40 repeat family protein] |
0.514 |
0.322 |
| 31 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.488 |
0.148 |
| 32 |
250796_at |
AT5G05300
|
unknown |
0.487 |
0.256 |
| 33 |
263475_at |
AT2G31945
|
unknown |
0.485 |
0.317 |
| 34 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
0.483 |
0.096 |
| 35 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
0.465 |
0.540 |
| 36 |
263182_at |
AT1G05575
|
unknown |
0.461 |
1.023 |
| 37 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.461 |
0.005 |
| 38 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
0.450 |
-0.312 |
| 39 |
258707_at |
AT3G09480
|
[Histone superfamily protein] |
0.449 |
0.446 |
| 40 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.449 |
0.026 |
| 41 |
246584_at |
AT5G14730
|
unknown |
0.446 |
0.151 |
| 42 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.446 |
0.229 |
| 43 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
0.434 |
0.287 |
| 44 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
0.432 |
0.166 |
| 45 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.425 |
0.898 |
| 46 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.422 |
0.529 |
| 47 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.403 |
0.643 |
| 48 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.402 |
0.176 |
| 49 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.399 |
1.111 |
| 50 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
0.396 |
0.228 |
| 51 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.394 |
0.112 |
| 52 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.391 |
0.429 |
| 53 |
266296_at |
AT2G29420
|
ATGSTU7, glutathione S-transferase tau 7, GST25, GLUTATHIONE S-TRANSFERASE 25, GSTU7, glutathione S-transferase tau 7 |
0.389 |
-0.070 |
| 54 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.388 |
0.068 |
| 55 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
0.385 |
0.170 |
| 56 |
257670_at |
AT3G20340
|
unknown |
0.383 |
-0.037 |
| 57 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.380 |
0.637 |
| 58 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
0.377 |
0.266 |
| 59 |
252825_at |
AT4G39890
|
AtRABH1c, RAB GTPase homolog H1C, RABH1c, RAB GTPase homolog H1C |
0.370 |
0.279 |
| 60 |
248821_at |
AT5G47070
|
[Protein kinase superfamily protein] |
0.369 |
0.180 |
| 61 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.407 |
-0.176 |
| 62 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
-0.386 |
-0.063 |
| 63 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
-0.354 |
-0.355 |
| 64 |
259994_at |
AT1G68130
|
AtIDD14, indeterminate(ID)-domain 14, IDD14, indeterminate(ID)-domain 14, IDD14alpha, IDD14beta |
-0.334 |
-0.147 |
| 65 |
254631_at |
AT4G18610
|
LSH9, LIGHT SENSITIVE HYPOCOTYLS 9 |
-0.317 |
-0.003 |
| 66 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
-0.309 |
-0.100 |
| 67 |
261500_at |
AT1G28400
|
unknown |
-0.294 |
-0.035 |
| 68 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
-0.294 |
-0.128 |
| 69 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.288 |
-0.172 |
| 70 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
-0.284 |
-0.157 |
| 71 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.282 |
-0.063 |
| 72 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
-0.280 |
-0.237 |
| 73 |
266797_at |
AT2G22840
|
AtGRF1, growth-regulating factor 1, GRF1, growth-regulating factor 1 |
-0.277 |
0.265 |
| 74 |
264455_at |
AT1G10330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.276 |
-0.142 |
| 75 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.271 |
-0.111 |
| 76 |
262349_at |
AT2G48130
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.263 |
-0.138 |
| 77 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.261 |
0.168 |
| 78 |
266824_at |
AT2G22800
|
HAT9 |
-0.253 |
-0.085 |
| 79 |
261451_at |
AT1G21060
|
unknown |
-0.252 |
-0.226 |
| 80 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.243 |
-0.171 |
| 81 |
254834_at |
AT4G12300
|
CYP706A4, cytochrome P450, family 706, subfamily A, polypeptide 4 |
-0.243 |
-0.188 |
| 82 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.235 |
-0.158 |
| 83 |
263738_at |
AT1G60060
|
[Serine/threonine-protein kinase WNK (With No Lysine)-related] |
-0.233 |
0.010 |
| 84 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.233 |
-0.345 |
| 85 |
261146_at |
AT1G19620
|
unknown |
-0.228 |
-0.091 |
| 86 |
262096_at |
AT1G56010
|
anac021, Arabidopsis NAC domain containing protein 21, ANAC022, Arabidopsis NAC domain containing protein 22, NAC1, NAC domain containing protein 1 |
-0.228 |
-0.286 |
| 87 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.227 |
0.535 |
| 88 |
245933_at |
AT5G09320
|
VPS9B |
-0.221 |
-0.107 |
| 89 |
246891_at |
AT5G25490
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.220 |
-0.080 |
| 90 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
-0.220 |
-0.261 |
| 91 |
261491_at |
AT1G14350
|
AtMYB124, myb domain protein 124, FLP, FOUR LIPS, MYB124 |
-0.218 |
0.056 |
| 92 |
249986_at |
AT5G18460
|
unknown |
-0.217 |
-0.119 |
| 93 |
255596_at |
AT4G01720
|
AtWRKY47, WRKY47 |
-0.210 |
-0.059 |
| 94 |
258362_at |
AT3G14280
|
unknown |
-0.209 |
-0.236 |
| 95 |
261203_at |
AT1G12845
|
unknown |
-0.208 |
0.257 |
| 96 |
261012_at |
AT1G26600
|
CLE9, CLAVATA3/ESR-RELATED 9 |
-0.207 |
-0.015 |
| 97 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.199 |
0.564 |
| 98 |
253746_at |
AT4G29100
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.198 |
-0.151 |
| 99 |
247087_at |
AT5G66330
|
[Leucine-rich repeat (LRR) family protein] |
-0.195 |
-0.037 |
| 100 |
247628_at |
AT5G60400
|
unknown |
-0.192 |
0.043 |
| 101 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.188 |
0.132 |
| 102 |
249779_at |
AT5G24230
|
[Lipase class 3-related protein] |
-0.184 |
-0.198 |
| 103 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.183 |
-0.097 |
| 104 |
247992_at |
AT5G56520
|
unknown |
-0.182 |
-0.095 |
| 105 |
253822_at |
AT4G28410
|
RSA1, Root System Architecture 1 |
-0.179 |
-0.113 |
| 106 |
257134_at |
AT3G12870
|
unknown |
-0.178 |
0.337 |
| 107 |
262286_at |
AT1G68585
|
unknown |
-0.178 |
-0.103 |
| 108 |
248715_at |
AT5G48290
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.177 |
-0.244 |
| 109 |
259240_at |
AT3G11590
|
unknown |
-0.176 |
0.025 |
| 110 |
255015_at |
AT4G09980
|
EMB1691, EMBRYO DEFECTIVE 1691 |
-0.176 |
-0.069 |
| 111 |
256395_at |
AT3G06120
|
MUTE, MUTE |
-0.176 |
0.094 |
| 112 |
266229_at |
AT2G28840
|
XBAT31, XB3 ortholog 1 in Arabidopsis thaliana |
-0.172 |
0.051 |
| 113 |
254561_at |
AT4G19160
|
unknown |
-0.168 |
-0.113 |
| 114 |
264987_at |
AT1G27030
|
unknown |
-0.168 |
0.202 |
| 115 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-0.167 |
-0.042 |
| 116 |
253500_at |
AT4G31920
|
ARR10, response regulator 10, RR10, response regulator 10 |
-0.167 |
-0.122 |
| 117 |
248908_at |
AT5G45800
|
MEE62, maternal effect embryo arrest 62 |
-0.166 |
-0.154 |
| 118 |
251744_at |
AT3G56010
|
unknown |
-0.166 |
-0.112 |
| 119 |
253525_at |
AT4G31330
|
unknown |
-0.163 |
-0.245 |