|
probeID |
AGICode |
Annotation |
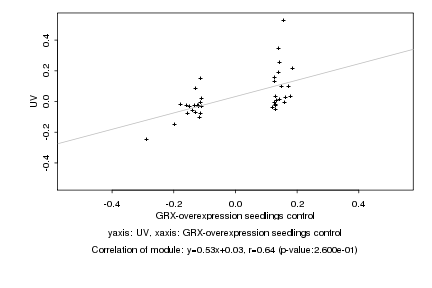
Log2 signal ratio
GRX-overexpression seedlings control |
Log2 signal ratio
UV |
| 1 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
0.184 |
0.218 |
| 2 |
262396_at |
AT1G49470
|
unknown |
0.178 |
0.037 |
| 3 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
0.169 |
0.099 |
| 4 |
260703_at |
AT1G32270
|
ATSYP24, SYP24, SYNTAXIN 24 |
0.160 |
0.028 |
| 5 |
265180_at |
AT1G23590
|
unknown |
0.159 |
-0.002 |
| 6 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.154 |
0.534 |
| 7 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.149 |
0.100 |
| 8 |
249576_at |
AT5G37690
|
[SGNH hydrolase-type esterase superfamily protein] |
0.142 |
0.014 |
| 9 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
0.141 |
0.259 |
| 10 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.139 |
0.349 |
| 11 |
255342_at |
AT4G04510
|
CRK38, cysteine-rich RLK (RECEPTOR-like protein kinase) 38 |
0.139 |
0.195 |
| 12 |
265758_at |
AT2G13070
|
unknown |
0.132 |
0.013 |
| 13 |
252054_at |
AT3G52540
|
ATOFP18, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 18, OFP18, ovate family protein 18 |
0.129 |
-0.049 |
| 14 |
253187_at |
AT4G35280
|
DAZ2, DUO1-ACTIVATED ZINC FINGER 2 |
0.128 |
-0.018 |
| 15 |
262102_at |
AT1G02980
|
ATCUL2, CUL2, cullin 2 |
0.127 |
0.035 |
| 16 |
251003_at |
AT5G02690
|
unknown |
0.127 |
-0.025 |
| 17 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.126 |
0.135 |
| 18 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.125 |
0.159 |
| 19 |
256604_at |
AT3G29640
|
unknown |
0.124 |
-0.005 |
| 20 |
250519_at |
AT5G08460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.119 |
-0.038 |
| 21 |
244994_at |
ATCG01010
|
NDHF |
-0.290 |
-0.245 |
| 22 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.200 |
-0.145 |
| 23 |
265363_at |
AT2G13335
|
[gypsy-like retrotransposon family, has a 9.7e-06 P-value blast match to GB:BAA84457 GAG-POL precursor (gypsy_Ty3-element) (Oryza sativa)gi|5902444|dbj|BAA84457.1| GAG-POL precursor (Oryza sativa (japonica cultivar-group)) (RIRE2) (Gypsy_Ty3-family)] |
-0.181 |
-0.019 |
| 24 |
259201_at |
AT3G09080
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.160 |
-0.020 |
| 25 |
252128_at |
AT3G50870
|
GATA18, GATA TRANSCRIPTION FACTOR 18, HAN, HANABA TANARU, MNP, MONOPOLE |
-0.156 |
-0.077 |
| 26 |
252742_at |
AT3G43290
|
unknown |
-0.151 |
-0.031 |
| 27 |
262211_at |
AT1G74930
|
ORA47 |
-0.140 |
-0.058 |
| 28 |
254768_at |
AT4G13320
|
unknown |
-0.135 |
-0.021 |
| 29 |
251778_at |
AT3G55660
|
ATROPGEF6, ROPGEF6, ROP (rho of plants) guanine nucleotide exchange factor 6 |
-0.131 |
-0.070 |
| 30 |
245451_at |
AT4G16890
|
BAL, BALL, SNC1, SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 |
-0.130 |
0.090 |
| 31 |
255828_at |
AT2G40630
|
[Uncharacterised conserved protein (UCP030365)] |
-0.124 |
-0.024 |
| 32 |
260473_at |
AT1G10880
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.122 |
-0.021 |
| 33 |
247052_at |
AT5G66700
|
ATHB53, ARABIDOPSIS THALIANA HOMEOBOX 53, HB-8, HOMEOBOX-8, HB53, homeobox 53 |
-0.120 |
-0.032 |
| 34 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.117 |
-0.100 |
| 35 |
249028_at |
AT5G44740
|
POLH, Y-family DNA polymerase H |
-0.114 |
-0.072 |
| 36 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.114 |
0.152 |
| 37 |
255172_x_at |
AT4G08000
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 1.0e-136 P-value blast match to GB:CAA40555 TNP2 (CACTA-element) (Antirrhinum majus)] |
-0.114 |
-0.006 |
| 38 |
246329_at |
AT3G43610
|
[Spc97 / Spc98 family of spindle pole body (SBP) component] |
-0.112 |
-0.031 |
| 39 |
249511_at |
AT5G38500
|
[FUNCTIONS IN: DNA binding] |
-0.111 |
0.021 |