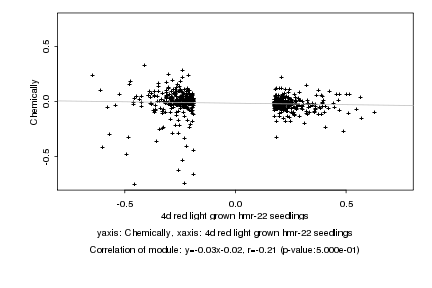
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
4d red light grown hmr-22 seedlings |
Log2 signal ratio
Chemically |
| 1 |
245233_at |
AT4G25580
|
[CAP160 protein] |
0.626 |
-0.092 |
| 2 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.568 |
-0.147 |
| 3 |
244999_at |
ATCG00190
|
RPOB, RNA polymerase subunit beta |
0.563 |
0.044 |
| 4 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
0.544 |
-0.067 |
| 5 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.511 |
0.069 |
| 6 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.507 |
-0.103 |
| 7 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
0.498 |
0.067 |
| 8 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.485 |
-0.271 |
| 9 |
264815_at |
AT1G03620
|
[ELMO/CED-12 family protein] |
0.466 |
0.071 |
| 10 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.466 |
-0.073 |
| 11 |
253322_at |
AT4G33980
|
unknown |
0.465 |
0.014 |
| 12 |
244938_at |
ATCG01120
|
RPS15, chloroplast ribosomal protein S15 |
0.456 |
0.072 |
| 13 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.444 |
-0.051 |
| 14 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.439 |
-0.009 |
| 15 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
0.424 |
0.093 |
| 16 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.418 |
-0.055 |
| 17 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.405 |
-0.229 |
| 18 |
262040_at |
AT1G80080
|
AtRLP17, Receptor Like Protein 17, TMM, TOO MANY MOUTHS |
0.403 |
-0.041 |
| 19 |
254634_at |
AT4G18650
|
[transcription factor-related] |
0.398 |
-0.099 |
| 20 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.396 |
0.018 |
| 21 |
260744_at |
AT1G15010
|
unknown |
0.393 |
-0.015 |
| 22 |
250873_at |
AT5G03980
|
[SGNH hydrolase-type esterase superfamily protein] |
0.393 |
0.051 |
| 23 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.391 |
-0.045 |
| 24 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
0.389 |
-0.008 |
| 25 |
244997_at |
ATCG00170
|
RPOC2 |
0.388 |
0.063 |
| 26 |
260668_at |
AT1G19530
|
unknown |
0.387 |
-0.109 |
| 27 |
263881_at |
AT2G21820
|
unknown |
0.381 |
-0.060 |
| 28 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.379 |
-0.050 |
| 29 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.375 |
0.104 |
| 30 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
0.374 |
-0.117 |
| 31 |
244998_at |
ATCG00180
|
RPOC1 |
0.363 |
0.056 |
| 32 |
255285_at |
AT4G04630
|
unknown |
0.360 |
-0.042 |
| 33 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.360 |
-0.024 |
| 34 |
264774_at |
AT1G22890
|
unknown |
0.360 |
-0.072 |
| 35 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.359 |
-0.063 |
| 36 |
266518_at |
AT2G35170
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.357 |
-0.095 |
| 37 |
256970_at |
AT3G21090
|
ABCG15, ATP-binding cassette G15 |
0.356 |
-0.083 |
| 38 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
0.348 |
-0.034 |
| 39 |
265296_at |
AT2G14060
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.346 |
-0.013 |
| 40 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.336 |
-0.042 |
| 41 |
252539_at |
AT3G45730
|
unknown |
0.332 |
-0.096 |
| 42 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.330 |
-0.023 |
| 43 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
0.329 |
-0.052 |
| 44 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.329 |
-0.014 |
| 45 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.325 |
-0.117 |
| 46 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.324 |
-0.012 |
| 47 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
0.322 |
0.011 |
| 48 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
0.320 |
-0.084 |
| 49 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.320 |
0.146 |
| 50 |
259982_at |
AT1G76410
|
ATL8 |
0.317 |
-0.100 |
| 51 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.312 |
-0.198 |
| 52 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.307 |
-0.018 |
| 53 |
255733_at |
AT1G25400
|
unknown |
0.301 |
-0.068 |
| 54 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
0.301 |
0.006 |
| 55 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
0.300 |
-0.037 |
| 56 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
0.296 |
-0.043 |
| 57 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.294 |
-0.061 |
| 58 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.293 |
0.010 |
| 59 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.288 |
0.088 |
| 60 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.287 |
-0.131 |
| 61 |
267041_at |
AT2G34315
|
[Avirulence induced gene (AIG1) family protein] |
0.285 |
0.015 |
| 62 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.285 |
-0.028 |
| 63 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
0.282 |
-0.033 |
| 64 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.281 |
0.029 |
| 65 |
257850_at |
AT3G13065
|
SRF4, STRUBBELIG-receptor family 4 |
0.277 |
-0.041 |
| 66 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.274 |
-0.021 |
| 67 |
265070_at |
AT1G55510
|
BCDH BETA1, branched-chain alpha-keto acid decarboxylase E1 beta subunit |
0.274 |
-0.060 |
| 68 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.273 |
-0.072 |
| 69 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.272 |
0.004 |
| 70 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.271 |
-0.009 |
| 71 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.269 |
0.011 |
| 72 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.268 |
0.045 |
| 73 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.267 |
0.008 |
| 74 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
0.267 |
-0.056 |
| 75 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.266 |
-0.123 |
| 76 |
263118_at |
AT1G03090
|
MCCA |
0.266 |
-0.052 |
| 77 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.265 |
0.072 |
| 78 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.264 |
-0.117 |
| 79 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.262 |
-0.096 |
| 80 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
0.261 |
-0.052 |
| 81 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.259 |
-0.095 |
| 82 |
263475_at |
AT2G31945
|
unknown |
0.255 |
-0.004 |
| 83 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.254 |
-0.102 |
| 84 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.254 |
-0.021 |
| 85 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.253 |
-0.099 |
| 86 |
254200_at |
AT4G24110
|
unknown |
0.253 |
0.019 |
| 87 |
248585_at |
AT5G49640
|
unknown |
0.252 |
-0.024 |
| 88 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.250 |
-0.057 |
| 89 |
264821_at |
AT1G03470
|
NET3A, Networked 3A |
0.249 |
0.011 |
| 90 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.249 |
-0.007 |
| 91 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.249 |
-0.097 |
| 92 |
251166_at |
AT3G63350
|
AT-HSFA7B, HSFA7B, HEAT SHOCK TRANSCRIPTION FACTOR A7B |
0.247 |
0.026 |
| 93 |
249058_at |
AT5G44510
|
TAO1, target of AVRB operation1 |
0.246 |
-0.033 |
| 94 |
244932_at |
ATCG01060
|
PSAC |
0.246 |
0.004 |
| 95 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
0.245 |
-0.179 |
| 96 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.245 |
-0.024 |
| 97 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
0.243 |
-0.038 |
| 98 |
250616_at |
AT5G07280
|
EMS1, EXCESS MICROSPOROCYTES1, EXS, EXTRA SPOROGENOUS CELLS |
0.243 |
-0.004 |
| 99 |
261205_at |
AT1G12790
|
unknown |
0.243 |
-0.027 |
| 100 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
0.241 |
0.114 |
| 101 |
261673_at |
AT1G18280
|
LTPG3, glycosylphosphatidylinositol-anchored lipid protein transfer 3 |
0.241 |
-0.143 |
| 102 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
0.240 |
-0.015 |
| 103 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.239 |
0.038 |
| 104 |
245008_at |
ATCG00360
|
YCF3 |
0.239 |
0.089 |
| 105 |
261203_at |
AT1G12845
|
unknown |
0.238 |
-0.005 |
| 106 |
248713_at |
AT5G48180
|
AtNSP5, NSP5, nitrile specifier protein 5 |
0.238 |
0.033 |
| 107 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
0.238 |
-0.034 |
| 108 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
0.237 |
0.041 |
| 109 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.237 |
-0.050 |
| 110 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.237 |
0.009 |
| 111 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
0.236 |
-0.038 |
| 112 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
0.236 |
-0.010 |
| 113 |
258326_at |
AT3G22760
|
SOL1 |
0.236 |
-0.011 |
| 114 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
0.236 |
-0.042 |
| 115 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
0.235 |
-0.003 |
| 116 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.234 |
-0.130 |
| 117 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
0.234 |
-0.060 |
| 118 |
250809_at |
AT5G05140
|
[Transcription elongation factor (TFIIS) family protein] |
0.232 |
-0.012 |
| 119 |
258527_at |
AT3G06850
|
BCE2, DIN3, DARK INDUCIBLE 3, LTA1 |
0.232 |
-0.085 |
| 120 |
256061_at |
AT1G07040
|
unknown |
0.232 |
0.005 |
| 121 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.231 |
-0.063 |
| 122 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.229 |
0.000 |
| 123 |
266054_at |
AT2G40640
|
[RING/U-box superfamily protein] |
0.227 |
0.054 |
| 124 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.227 |
-0.090 |
| 125 |
263265_at |
AT2G38820
|
unknown |
0.227 |
-0.006 |
| 126 |
245781_at |
AT1G45976
|
SBP1, S-ribonuclease binding protein 1 |
0.226 |
-0.025 |
| 127 |
262977_at |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
0.225 |
-0.010 |
| 128 |
260837_at |
AT1G43670
|
AtcFBP, Arabidopsis thaliana cytosolic fructose-1,6-bisphosphatase, FBP, fructose-1,6-bisphosphatase, FINS1, FRUCTOSE INSENSITIVE 1 |
0.225 |
-0.011 |
| 129 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.224 |
0.110 |
| 130 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.224 |
0.011 |
| 131 |
261567_at |
AT1G33055
|
unknown |
0.224 |
-0.072 |
| 132 |
263281_at |
AT2G14160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.224 |
-0.173 |
| 133 |
262359_at |
AT1G73066
|
[Leucine-rich repeat family protein] |
0.224 |
-0.027 |
| 134 |
267375_at |
AT2G26300
|
ATGPA1, ARABIDOPSIS THALIANA G PROTEIN ALPHA SUBUNIT 1, GP ALPHA 1, G protein alpha subunit 1, GPA1, G PROTEIN ALPHA SUBUNIT 1 |
0.224 |
0.042 |
| 135 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.222 |
-0.061 |
| 136 |
260264_at |
AT1G68500
|
unknown |
0.220 |
-0.027 |
| 137 |
255881_at |
AT1G67070
|
DIN9, DARK INDUCIBLE 9, PMI2, PHOSPHOMANNOSE ISOMERASE 2 |
0.220 |
-0.058 |
| 138 |
267557_at |
AT2G32710
|
ACK2, CYCLIN-DEPENDENT KINASE INHIBITOR 2, ICK7, INTERACTORS OF CDC2 KINASE 7, KRP4, KRP4, KIP-RELATED PROTEIN 4 |
0.219 |
-0.051 |
| 139 |
264933_at |
AT1G61160
|
unknown |
0.219 |
0.075 |
| 140 |
250025_at |
AT5G18290
|
SIP1;2, SIP1B, SMALL AND BASIC INTRINSIC PROTEIN 1B |
0.219 |
-0.028 |
| 141 |
247599_at |
AT5G60880
|
BASL, BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE |
0.219 |
0.044 |
| 142 |
264236_at |
AT1G54680
|
unknown |
0.218 |
-0.065 |
| 143 |
258248_at |
AT3G29140
|
unknown |
0.217 |
0.044 |
| 144 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.217 |
-0.037 |
| 145 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.217 |
-0.028 |
| 146 |
251291_at |
AT3G61900
|
SAUR33, SMALL AUXIN UPREGULATED RNA 33 |
0.216 |
-0.070 |
| 147 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
0.216 |
0.001 |
| 148 |
261502_at |
AT1G14440
|
AtHB31, homeobox protein 31, FTM2, FLORAL TRANSITION AT THE MERISTEM2, HB31, homeobox protein 31, ZHD4, ZINC FINGER HOMEODOMAIN 4 |
0.216 |
0.035 |
| 149 |
248205_at |
AT5G54300
|
unknown |
0.216 |
-0.149 |
| 150 |
260272_at |
AT1G80570
|
[RNI-like superfamily protein] |
0.215 |
-0.057 |
| 151 |
264238_at |
AT1G54740
|
unknown |
0.214 |
-0.064 |
| 152 |
248204_at |
AT5G54280
|
ATM2, myosin 2, ATM4, ARABIDOPSIS THALIANA MYOSIN 4, ATMYOS1, ARABIDOPSIS THALIANA MYOSIN 1 |
0.214 |
0.010 |
| 153 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
0.214 |
-0.022 |
| 154 |
245314_at |
AT4G16745
|
[Exostosin family protein] |
0.214 |
-0.073 |
| 155 |
262205_at |
AT2G01080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.213 |
-0.002 |
| 156 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.213 |
-0.022 |
| 157 |
260536_at |
AT2G43400
|
ETFQO, electron-transfer flavoprotein:ubiquinone oxidoreductase |
0.213 |
-0.037 |
| 158 |
245703_at |
AT5G04380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.213 |
0.028 |
| 159 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.213 |
-0.042 |
| 160 |
245018_at |
ATCG00520
|
YCF4 |
0.212 |
0.071 |
| 161 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.212 |
-0.000 |
| 162 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.211 |
-0.023 |
| 163 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.211 |
0.002 |
| 164 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
0.211 |
0.013 |
| 165 |
246887_at |
AT5G26250
|
[Major facilitator superfamily protein] |
0.210 |
-0.046 |
| 166 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.210 |
-0.035 |
| 167 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.209 |
0.062 |
| 168 |
258581_at |
AT3G04160
|
unknown |
0.209 |
-0.008 |
| 169 |
247710_at |
AT5G59260
|
LecRK-II.1, L-type lectin receptor kinase II.1 |
0.208 |
-0.026 |
| 170 |
260279_at |
AT1G80420
|
ATXRCC1, XRCC1, homolog of X-ray repair cross complementing 1 |
0.208 |
0.021 |
| 171 |
251750_at |
AT3G55710
|
[UDP-Glycosyltransferase superfamily protein] |
0.208 |
-0.038 |
| 172 |
244939_at |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
0.208 |
0.224 |
| 173 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.207 |
-0.099 |
| 174 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.207 |
-0.012 |
| 175 |
262333_at |
AT1G64020
|
[Serine protease inhibitor (SERPIN) family protein] |
0.207 |
-0.008 |
| 176 |
264405_at |
AT1G10210
|
ATMPK1, mitogen-activated protein kinase 1, MPK1, mitogen-activated protein kinase 1 |
0.207 |
-0.074 |
| 177 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
0.206 |
0.051 |
| 178 |
256413_at |
AT3G11100
|
[sequence-specific DNA binding transcription factors] |
0.206 |
-0.013 |
| 179 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.206 |
-0.049 |
| 180 |
264296_at |
AT1G78720
|
[SecY protein transport family protein] |
0.206 |
-0.032 |
| 181 |
247616_at |
AT5G60260
|
unknown |
0.205 |
0.027 |
| 182 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
0.205 |
-0.017 |
| 183 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.205 |
0.039 |
| 184 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
0.204 |
-0.012 |
| 185 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.204 |
0.122 |
| 186 |
258472_at |
AT3G06080
|
TBL10, TRICHOME BIREFRINGENCE-LIKE 10 |
0.204 |
-0.045 |
| 187 |
264588_at |
AT2G17730
|
NIP2, NEP-interacting protein 2 |
0.203 |
-0.058 |
| 188 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.203 |
0.015 |
| 189 |
263702_at |
AT1G31240
|
[Bromodomain transcription factor] |
0.202 |
-0.013 |
| 190 |
255472_at |
AT4G02430
|
At-SR34b, Serine/Arginine-Rich Protein Splicing Factor 34b, SR34b, Serine/Arginine-Rich Protein Splicing Factor 34b |
0.202 |
0.012 |
| 191 |
261252_at |
AT1G05810
|
ARA, ARA-1, ATRAB11D, ATRABA5E, ARABIDOPSIS THALIANA RAB GTPASE HOMOLOG A5E, RABA5E, RAB GTPase homolog A5E |
0.201 |
-0.070 |
| 192 |
250766_at |
AT5G05550
|
[sequence-specific DNA binding transcription factors] |
0.200 |
0.024 |
| 193 |
260732_at |
AT1G17520
|
[Homeodomain-like/winged-helix DNA-binding family protein] |
0.200 |
-0.053 |
| 194 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
0.200 |
-0.052 |
| 195 |
259979_at |
AT1G76600
|
unknown |
0.199 |
-0.061 |
| 196 |
247979_at |
AT5G56750
|
NDL1, N-MYC downregulated-like 1 |
0.199 |
-0.054 |
| 197 |
258939_at |
AT3G10020
|
unknown |
0.199 |
-0.010 |
| 198 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.198 |
-0.135 |
| 199 |
253279_at |
AT4G34030
|
MCCB, 3-methylcrotonyl-CoA carboxylase |
0.198 |
-0.030 |
| 200 |
251022_at |
AT5G02150
|
Fes1C, Fes1C |
0.197 |
-0.009 |
| 201 |
264822_at |
AT1G03457
|
AtBRN2, BRN2, Bruno-like 2 |
0.197 |
-0.100 |
| 202 |
264362_at |
AT1G03290
|
unknown |
0.197 |
-0.012 |
| 203 |
257737_at |
AT3G27440
|
UKL5, uridine kinase-like 5 |
0.197 |
0.017 |
| 204 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.197 |
0.120 |
| 205 |
249772_at |
AT5G24130
|
unknown |
0.196 |
0.016 |
| 206 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.196 |
0.001 |
| 207 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.196 |
-0.023 |
| 208 |
266910_at |
AT2G45920
|
[U-box domain-containing protein] |
0.195 |
0.023 |
| 209 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.195 |
-0.018 |
| 210 |
256338_at |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.194 |
0.007 |
| 211 |
253406_at |
AT4G32890
|
GATA9, GATA transcription factor 9 |
0.194 |
0.062 |
| 212 |
262443_at |
AT1G47655
|
[Dof-type zinc finger DNA-binding family protein] |
0.194 |
0.011 |
| 213 |
247833_at |
AT5G58575
|
unknown |
0.193 |
0.007 |
| 214 |
257580_at |
AT3G06210
|
[ARM repeat superfamily protein] |
0.193 |
-0.005 |
| 215 |
262845_at |
AT1G14740
|
TTA1, TITANIA 1 |
0.193 |
0.027 |
| 216 |
251490_at |
AT3G59490
|
unknown |
0.193 |
-0.027 |
| 217 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.192 |
-0.035 |
| 218 |
252978_at |
AT4G38590
|
BGAL14, beta-galactosidase 14 |
0.192 |
0.030 |
| 219 |
249345_at |
AT5G40740
|
AUG6, augmin subunit 6 |
0.192 |
-0.055 |
| 220 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.191 |
0.010 |
| 221 |
254561_at |
AT4G19160
|
unknown |
0.191 |
-0.045 |
| 222 |
258191_at |
AT3G29100
|
ATVTI13, VTI13, vesicle transport V-snare 13 |
0.190 |
0.034 |
| 223 |
261674_at |
AT1G18270
|
[ketose-bisphosphate aldolase class-II family protein] |
0.190 |
-0.065 |
| 224 |
251421_at |
AT3G60510
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.190 |
-0.013 |
| 225 |
256457_at |
AT1G75230
|
[DNA glycosylase superfamily protein] |
0.189 |
-0.008 |
| 226 |
253224_at |
AT4G34860
|
A/N-InvB, alkaline/neutral invertase B |
0.189 |
0.008 |
| 227 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
0.188 |
0.002 |
| 228 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.188 |
-0.030 |
| 229 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.188 |
0.006 |
| 230 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.188 |
-0.012 |
| 231 |
252095_at |
AT3G51000
|
[alpha/beta-Hydrolases superfamily protein] |
0.187 |
-0.018 |
| 232 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.187 |
-0.044 |
| 233 |
246763_at |
AT5G27150
|
AT-NHX1, ATNHX, ATNHX1, NHX1, Na+/H+ exchanger 1 |
0.187 |
-0.021 |
| 234 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.187 |
0.005 |
| 235 |
263827_at |
AT2G40420
|
[Transmembrane amino acid transporter family protein] |
0.187 |
-0.059 |
| 236 |
252424_at |
AT3G47610
|
[transcription regulators] |
0.186 |
-0.032 |
| 237 |
247670_at |
AT5G60190
|
[Cysteine proteinases superfamily protein] |
0.186 |
-0.069 |
| 238 |
261030_at |
AT1G17410
|
[Nucleoside diphosphate kinase family protein] |
0.186 |
-0.041 |
| 239 |
260858_at |
AT1G43770
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.186 |
0.016 |
| 240 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
0.185 |
0.055 |
| 241 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.185 |
-0.003 |
| 242 |
247774_at |
AT5G58660
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.185 |
0.024 |
| 243 |
266593_at |
AT2G46200
|
unknown |
0.185 |
0.024 |
| 244 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.185 |
-0.038 |
| 245 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.184 |
-0.046 |
| 246 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
0.184 |
0.008 |
| 247 |
263275_at |
AT2G14170
|
ALDH6B2, aldehyde dehydrogenase 6B2 |
0.184 |
-0.037 |
| 248 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
0.184 |
-0.175 |
| 249 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.184 |
-0.008 |
| 250 |
245335_at |
AT4G16160
|
ATOEP16-2, ATOEP16-S |
0.183 |
0.042 |
| 251 |
252820_at |
AT3G42640
|
AHA8, H(+)-ATPase 8, HA8, H(+)-ATPase 8 |
0.183 |
-0.062 |
| 252 |
266929_at |
AT2G45850
|
AHL9, AT-hook motif nuclear localized protein 9 |
0.183 |
-0.006 |
| 253 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.182 |
0.004 |
| 254 |
248969_at |
AT5G45310
|
unknown |
0.182 |
0.008 |
| 255 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.182 |
0.122 |
| 256 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
0.182 |
-0.049 |
| 257 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.182 |
-0.048 |
| 258 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
0.182 |
0.061 |
| 259 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.181 |
-0.325 |
| 260 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.181 |
-0.043 |
| 261 |
265993_at |
AT2G24160
|
[pseudogene] |
0.181 |
0.010 |
| 262 |
245132_at |
AT2G45320
|
unknown |
0.181 |
-0.020 |
| 263 |
265989_at |
AT2G24260
|
LRL1, LJRHL1-like 1 |
0.181 |
-0.065 |
| 264 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
0.180 |
0.117 |
| 265 |
259984_at |
AT1G76460
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.180 |
-0.062 |
| 266 |
256544_at |
AT1G42560
|
ATMLO9, ARABIDOPSIS THALIANA MILDEW RESISTANCE LOCUS O 9, MLO9, MILDEW RESISTANCE LOCUS O 9 |
0.180 |
-0.066 |
| 267 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.180 |
0.019 |
| 268 |
263113_at |
AT1G03150
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.180 |
-0.012 |
| 269 |
267054_at |
AT2G38370
|
unknown |
0.180 |
0.049 |
| 270 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.180 |
-0.012 |
| 271 |
267268_at |
AT2G02570
|
[nucleic acid binding] |
0.180 |
0.007 |
| 272 |
253126_at |
AT4G36050
|
[endonuclease/exonuclease/phosphatase family protein] |
0.180 |
-0.017 |
| 273 |
251641_at |
AT3G57470
|
[Insulinase (Peptidase family M16) family protein] |
0.179 |
0.001 |
| 274 |
262487_at |
AT1G21610
|
[wound-responsive family protein] |
0.178 |
-0.029 |
| 275 |
253096_at |
AT4G37330
|
CYP81D4, cytochrome P450, family 81, subfamily D, polypeptide 4 |
0.178 |
-0.046 |
| 276 |
250860_at |
AT5G04770
|
ATCAT6, ARABIDOPSIS THALIANA CATIONIC AMINO ACID TRANSPORTER 6, CAT6, cationic amino acid transporter 6 |
0.178 |
-0.047 |
| 277 |
261437_at |
AT1G07745
|
ATRAD51D, RAD51D, homolog of RAD51 D, SSN1, SUPPRESOR OF SNI1 |
0.178 |
-0.018 |
| 278 |
262295_at |
AT1G27650
|
ATU2AF35A |
0.178 |
0.004 |
| 279 |
258501_at |
AT3G06780
|
[glycine-rich protein] |
0.178 |
-0.036 |
| 280 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.178 |
-0.049 |
| 281 |
251138_at |
AT5G01160
|
[RING/U-box superfamily protein] |
0.178 |
0.007 |
| 282 |
252210_at |
AT3G50410
|
OBP1, OBF binding protein 1 |
0.178 |
0.059 |
| 283 |
254787_at |
AT4G12690
|
unknown |
0.177 |
-0.072 |
| 284 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.177 |
-0.035 |
| 285 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
0.176 |
-0.024 |
| 286 |
259209_at |
AT3G09160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.176 |
-0.062 |
| 287 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.176 |
-0.024 |
| 288 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
0.176 |
-0.002 |
| 289 |
247394_at |
AT5G62865
|
unknown |
0.176 |
-0.073 |
| 290 |
262114_at |
AT1G02860
|
BAH1, BENZOIC ACID HYPERSENSITIVE 1, NLA, nitrogen limitation adaptation |
0.175 |
0.019 |
| 291 |
260627_at |
AT1G62310
|
[transcription factor jumonji (jmjC) domain-containing protein] |
0.175 |
-0.005 |
| 292 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.175 |
-0.017 |
| 293 |
256998_at |
AT3G14180
|
ASIL2, Arabidopsis 6B-interacting protein 1-like 2 |
0.175 |
-0.012 |
| 294 |
246881_at |
AT5G26040
|
HDA2, histone deacetylase 2 |
0.174 |
-0.034 |
| 295 |
253832_at |
AT4G27654
|
unknown |
0.174 |
-0.129 |
| 296 |
256149_at |
AT1G55110
|
AtIDD7, indeterminate(ID)-domain 7, IDD7, indeterminate(ID)-domain 7 |
0.174 |
-0.037 |
| 297 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
0.174 |
0.020 |
| 298 |
265495_at |
AT2G15695
|
unknown |
0.174 |
-0.030 |
| 299 |
254651_at |
AT4G18160
|
ATKCO6, CA2+ ACTIVATED OUTWARD RECTIFYING K+ CHANNEL 6, ATTPK3, KCO6, Ca2+ activated outward rectifying K+ channel 6, TPK3 |
0.173 |
-0.003 |
| 300 |
252100_at |
AT3G51110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.173 |
0.004 |
| 301 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.649 |
0.244 |
| 302 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.610 |
0.102 |
| 303 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.602 |
-0.412 |
| 304 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.579 |
-0.048 |
| 305 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.572 |
-0.291 |
| 306 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.544 |
-0.029 |
| 307 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.527 |
0.068 |
| 308 |
250778_at |
AT5G05500
|
MOP10 |
-0.495 |
-0.472 |
| 309 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.484 |
-0.322 |
| 310 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.481 |
0.156 |
| 311 |
258396_at |
AT3G15460
|
[Ribosomal RNA processing Brix domain protein] |
-0.477 |
0.188 |
| 312 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.464 |
-0.004 |
| 313 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.462 |
-0.018 |
| 314 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.460 |
-0.744 |
| 315 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.457 |
0.031 |
| 316 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.451 |
0.053 |
| 317 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.435 |
0.026 |
| 318 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.427 |
-0.045 |
| 319 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
-0.427 |
0.046 |
| 320 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
-0.425 |
-0.002 |
| 321 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.415 |
0.334 |
| 322 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.397 |
0.058 |
| 323 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.397 |
0.056 |
| 324 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
-0.389 |
0.093 |
| 325 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.384 |
0.038 |
| 326 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.384 |
-0.016 |
| 327 |
267376_at |
AT2G26330
|
ER, ERECTA, QRP1, QUANTITATIVE RESISTANCE TO PLECTOSPHAERELLA 1 |
-0.381 |
-0.024 |
| 328 |
251058_at |
AT5G01790
|
unknown |
-0.381 |
-0.010 |
| 329 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.380 |
0.076 |
| 330 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.373 |
-0.077 |
| 331 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
-0.369 |
0.052 |
| 332 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.369 |
-0.093 |
| 333 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.368 |
0.100 |
| 334 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.368 |
0.056 |
| 335 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.365 |
-0.077 |
| 336 |
265463_at |
AT2G37090
|
IRX9, IRREGULAR XYLEM 9 |
-0.364 |
-0.035 |
| 337 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.364 |
-0.068 |
| 338 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.357 |
-0.356 |
| 339 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.355 |
0.046 |
| 340 |
253233_at |
AT4G34290
|
[SWIB/MDM2 domain superfamily protein] |
-0.354 |
0.002 |
| 341 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.350 |
0.145 |
| 342 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.349 |
0.095 |
| 343 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.348 |
0.171 |
| 344 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.346 |
-0.246 |
| 345 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.346 |
0.003 |
| 346 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.342 |
-0.056 |
| 347 |
257173_at |
AT3G23810
|
ATSAHH2, S-ADENOSYL-L-HOMOCYSTEINE (SAH) HYDROLASE 2, SAHH2, S-adenosyl-l-homocysteine (SAH) hydrolase 2 |
-0.336 |
0.024 |
| 348 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.334 |
0.077 |
| 349 |
261684_at |
AT1G47400
|
unknown |
-0.332 |
-0.243 |
| 350 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-0.332 |
-0.108 |
| 351 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.332 |
-0.087 |
| 352 |
263488_at |
AT2G31840
|
MRL7-L, Mesophyll-cell RNAi Library line 7-like |
-0.328 |
0.001 |
| 353 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.327 |
-0.068 |
| 354 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.326 |
-0.227 |
| 355 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.324 |
0.004 |
| 356 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.320 |
-0.017 |
| 357 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
-0.317 |
-0.091 |
| 358 |
257608_at |
AT3G13860
|
HSP60-3A, heat shock protein 60-3A |
-0.316 |
0.034 |
| 359 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
-0.316 |
0.108 |
| 360 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.315 |
-0.003 |
| 361 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.315 |
0.046 |
| 362 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.315 |
0.090 |
| 363 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
-0.313 |
0.001 |
| 364 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.313 |
-0.003 |
| 365 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.313 |
0.041 |
| 366 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.312 |
0.039 |
| 367 |
255460_at |
AT4G02800
|
unknown |
-0.312 |
0.179 |
| 368 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
-0.312 |
0.060 |
| 369 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
-0.309 |
0.056 |
| 370 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.305 |
-0.018 |
| 371 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.304 |
-0.004 |
| 372 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
-0.304 |
0.130 |
| 373 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.304 |
0.251 |
| 374 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.303 |
0.122 |
| 375 |
261080_at |
AT1G07370
|
ATPCNA1, PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1, PCNA1, proliferating cellular nuclear antigen 1 |
-0.302 |
0.070 |
| 376 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.300 |
0.032 |
| 377 |
259964_at |
AT1G53680
|
ATGSTU28, glutathione S-transferase TAU 28, GSTU28, glutathione S-transferase TAU 28 |
-0.297 |
-0.157 |
| 378 |
250337_at |
AT5G11790
|
NDL2, N-MYC downregulated-like 2 |
-0.297 |
-0.017 |
| 379 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.296 |
0.033 |
| 380 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.296 |
-0.031 |
| 381 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
-0.296 |
-0.093 |
| 382 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.292 |
-0.058 |
| 383 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.292 |
0.019 |
| 384 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.292 |
0.003 |
| 385 |
251029_at |
AT5G02050
|
[Mitochondrial glycoprotein family protein] |
-0.291 |
0.053 |
| 386 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
-0.291 |
-0.029 |
| 387 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
-0.290 |
0.013 |
| 388 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.289 |
0.091 |
| 389 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.288 |
0.038 |
| 390 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.288 |
0.198 |
| 391 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.287 |
0.013 |
| 392 |
265049_at |
AT1G52060
|
[Mannose-binding lectin superfamily protein] |
-0.286 |
-0.282 |
| 393 |
266244_at |
AT2G27740
|
unknown |
-0.285 |
0.029 |
| 394 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.284 |
0.117 |
| 395 |
248074_at |
AT5G55730
|
FLA1, FASCICLIN-like arabinogalactan 1 |
-0.283 |
0.001 |
| 396 |
263882_at |
AT2G21790
|
ATRNR1, RIBONUCLEOTIDE REDUCTASE LARGE SUBUNIT 1, CLS8, CRINKLY LEAVES 8, DPD2, defective in pollen organelle DNA degradation 2, R1, RIBONUCLEOTIDE REDUCTASE 1, RNR1, ribonucleotide reductase 1 |
-0.283 |
0.079 |
| 397 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.282 |
-0.023 |
| 398 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.282 |
0.004 |
| 399 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-0.280 |
-0.052 |
| 400 |
248139_at |
AT5G54970
|
unknown |
-0.278 |
0.070 |
| 401 |
259678_at |
AT1G77750
|
[Ribosomal protein S13/S18 family] |
-0.277 |
0.000 |
| 402 |
247651_at |
AT5G59870
|
HTA6, histone H2A 6 |
-0.275 |
0.006 |
| 403 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.275 |
0.075 |
| 404 |
253654_at |
AT4G30060
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.275 |
-0.020 |
| 405 |
251857_at |
AT3G54770
|
ARP1, ABA-regulated RNA-binding protein 1 |
-0.274 |
-0.214 |
| 406 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.274 |
0.111 |
| 407 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
-0.273 |
0.071 |
| 408 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.273 |
-0.010 |
| 409 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
-0.273 |
0.082 |
| 410 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
-0.271 |
0.076 |
| 411 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.270 |
0.093 |
| 412 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
-0.270 |
-0.013 |
| 413 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
-0.270 |
0.129 |
| 414 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
-0.270 |
-0.118 |
| 415 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.270 |
0.050 |
| 416 |
248150_at |
AT5G54670
|
ATK3, kinesin 3, KATC, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA C |
-0.270 |
-0.094 |
| 417 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.270 |
0.054 |
| 418 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.269 |
0.096 |
| 419 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.269 |
-0.028 |
| 420 |
266264_at |
AT2G27775
|
unknown |
-0.268 |
0.022 |
| 421 |
249439_at |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.265 |
0.002 |
| 422 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.265 |
0.021 |
| 423 |
249167_at |
AT5G42860
|
unknown |
-0.264 |
-0.031 |
| 424 |
260028_at |
AT1G29980
|
unknown |
-0.263 |
0.162 |
| 425 |
248105_at |
AT5G55280
|
ATFTSZ1-1, ARABIDOPSIS THALIANA HOMOLOG OF BACTERIAL CYTOKINESIS Z-RING PROTEIN FTSZ 1-1, CPFTSZ, CHLOROPLAST FTSZ, FTSZ1-1, homolog of bacterial cytokinesis Z-ring protein FTSZ 1-1 |
-0.260 |
-0.006 |
| 426 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.259 |
-0.005 |
| 427 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.259 |
-0.284 |
| 428 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.259 |
-0.087 |
| 429 |
252077_at |
AT3G51720
|
unknown |
-0.258 |
0.098 |
| 430 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
-0.258 |
-0.170 |
| 431 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.258 |
-0.621 |
| 432 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.258 |
0.005 |
| 433 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.257 |
0.016 |
| 434 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
-0.256 |
0.094 |
| 435 |
261933_at |
AT1G22410
|
[Class-II DAHP synthetase family protein] |
-0.256 |
-0.002 |
| 436 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.256 |
-0.011 |
| 437 |
255005_at |
AT4G09990
|
GXM2, glucuronoxylan methyltransferase 2 |
-0.255 |
-0.212 |
| 438 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.255 |
0.005 |
| 439 |
247425_at |
AT5G62550
|
unknown |
-0.255 |
0.103 |
| 440 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
-0.255 |
0.138 |
| 441 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.254 |
-0.036 |
| 442 |
254629_at |
AT4G18425
|
unknown |
-0.254 |
-0.019 |
| 443 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.253 |
0.016 |
| 444 |
252089_at |
AT3G52110
|
unknown |
-0.253 |
0.026 |
| 445 |
267636_at |
AT2G42110
|
unknown |
-0.253 |
0.038 |
| 446 |
248121_at |
AT5G54690
|
GAUT12, galacturonosyltransferase 12, IRX8, IRREGULAR XYLEM 8, LGT6 |
-0.252 |
0.034 |
| 447 |
246425_at |
AT5G17420
|
ATCESA7, CESA7, CELLULOSE SYNTHASE CATALYTIC SUBUNIT 7, IRX3, IRREGULAR XYLEM 3, MUR10, MURUS 10 |
-0.252 |
-0.045 |
| 448 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
-0.252 |
-0.042 |
| 449 |
246415_at |
AT5G17160
|
unknown |
-0.252 |
0.190 |
| 450 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
-0.251 |
-0.025 |
| 451 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.251 |
0.112 |
| 452 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.251 |
0.025 |
| 453 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.250 |
-0.055 |
| 454 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.250 |
0.047 |
| 455 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
-0.249 |
0.014 |
| 456 |
259710_at |
AT1G77670
|
[Pyridoxal phosphate (PLP)-dependent transferases superfamily protein] |
-0.247 |
0.033 |
| 457 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.246 |
0.052 |
| 458 |
264969_at |
AT1G67320
|
EMB2813, EMBRYO DEFECTIVE 2813 |
-0.245 |
0.093 |
| 459 |
263845_at |
AT2G37040
|
ATPAL1, PAL1, PHE ammonia lyase 1 |
-0.245 |
0.024 |
| 460 |
249276_at |
AT5G41880
|
POLA3, POLA4 |
-0.244 |
0.042 |
| 461 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.243 |
0.019 |
| 462 |
245164_at |
AT2G33210
|
HSP60-2, heat shock protein 60-2 |
-0.243 |
0.036 |
| 463 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.243 |
0.284 |
| 464 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
-0.243 |
-0.009 |
| 465 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.242 |
0.087 |
| 466 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
-0.242 |
0.044 |
| 467 |
253777_at |
AT4G28450
|
[nucleotide binding] |
-0.242 |
0.059 |
| 468 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
-0.242 |
0.023 |
| 469 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.242 |
0.225 |
| 470 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.242 |
0.084 |
| 471 |
256905_at |
AT3G23990
|
HSP60, heat shock protein 60, HSP60-3B, HEAT SHOCK PROTEIN 60-3B |
-0.241 |
0.053 |
| 472 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.240 |
-0.534 |
| 473 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.240 |
0.052 |
| 474 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
-0.239 |
0.065 |
| 475 |
248085_at |
AT5G55480
|
GDPDL4, Glycerophosphodiester phosphodiesterase (GDPD) like 4, GPDL1, glycerophosphodiesterase-like 1, SVL1, SHV3-like 1 |
-0.239 |
-0.096 |
| 476 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
-0.239 |
-0.031 |
| 477 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
-0.239 |
-0.043 |
| 478 |
251093_at |
AT5G01360
|
TBL3, TRICHOME BIREFRINGENCE-LIKE 3 |
-0.238 |
-0.050 |
| 479 |
265961_at |
AT2G37400
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.237 |
0.001 |
| 480 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.237 |
-0.003 |
| 481 |
253090_at |
AT4G36360
|
BGAL3, beta-galactosidase 3 |
-0.236 |
0.081 |
| 482 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.235 |
0.006 |
| 483 |
261439_at |
AT1G28395
|
unknown |
-0.235 |
-0.000 |
| 484 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.234 |
-0.031 |
| 485 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.234 |
-0.738 |
| 486 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
-0.234 |
0.015 |
| 487 |
262094_at |
AT1G56110
|
NOP56, homolog of nucleolar protein NOP56 |
-0.234 |
0.113 |
| 488 |
266221_at |
AT2G28760
|
UXS6, UDP-XYL synthase 6 |
-0.234 |
-0.033 |
| 489 |
247068_at |
AT5G66800
|
unknown |
-0.233 |
-0.048 |
| 490 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.233 |
-0.130 |
| 491 |
247235_at |
AT5G64580
|
EMB3144, EMBRYO DEFECTIVE 3144 |
-0.233 |
-0.017 |
| 492 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
-0.233 |
0.011 |
| 493 |
252361_at |
AT3G48490
|
unknown |
-0.231 |
-0.007 |
| 494 |
256935_at |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.231 |
-0.330 |
| 495 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
-0.229 |
0.023 |
| 496 |
249759_at |
AT5G24380
|
ATYSL2, YELLOW STRIPE LIKE 2, YSL2, YELLOW STRIPE like 2 |
-0.229 |
-0.023 |
| 497 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
-0.228 |
0.011 |
| 498 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.228 |
-0.009 |
| 499 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
-0.228 |
-0.048 |
| 500 |
258633_at |
AT3G07990
|
SCPL27, serine carboxypeptidase-like 27 |
-0.227 |
0.016 |
| 501 |
267382_at |
AT2G44300
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.226 |
0.011 |
| 502 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-0.226 |
0.050 |
| 503 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.225 |
-0.046 |
| 504 |
245367_at |
AT4G16265
|
NRPB9B, NRPD9B, NRPE9B |
-0.225 |
0.051 |
| 505 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
-0.225 |
0.075 |
| 506 |
246511_at |
AT5G15490
|
UGD3, UDP-glucose dehydrogenase 3 |
-0.224 |
0.008 |
| 507 |
265960_at |
AT2G37470
|
[Histone superfamily protein] |
-0.224 |
-0.006 |
| 508 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
-0.224 |
0.016 |
| 509 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
-0.224 |
0.018 |
| 510 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-0.224 |
0.016 |
| 511 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.224 |
0.076 |
| 512 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
-0.223 |
-0.050 |
| 513 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.223 |
-0.039 |
| 514 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.223 |
0.097 |
| 515 |
260143_at |
AT1G71880
|
ATSUC1, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 1, SUC1, sucrose-proton symporter 1 |
-0.222 |
0.015 |
| 516 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
-0.222 |
0.032 |
| 517 |
265340_at |
AT2G18330
|
[AAA-type ATPase family protein] |
-0.222 |
0.011 |
| 518 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.222 |
-0.402 |
| 519 |
247268_at |
AT5G64080
|
AtXYP1, XYP1, xylogen protein 1 |
-0.221 |
0.072 |
| 520 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.221 |
0.014 |
| 521 |
260857_at |
AT1G21880
|
LYM1, lysm domain GPI-anchored protein 1 precursor, LYP2, LysM-containing receptor protein 2 |
-0.221 |
-0.005 |
| 522 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.220 |
-0.004 |
| 523 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.220 |
-0.004 |
| 524 |
248101_at |
AT5G55200
|
MGE1, mitochondrial GrpE 1 |
-0.219 |
0.015 |
| 525 |
245792_at |
AT1G32100
|
ATPRR1, PRR1, pinoresinol reductase 1 |
-0.219 |
0.003 |
| 526 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.219 |
-0.172 |
| 527 |
264493_at |
AT1G27440
|
ATGUT1, GUT2, IRX10 |
-0.218 |
-0.005 |
| 528 |
250230_at |
AT5G13900
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.217 |
-0.051 |
| 529 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.217 |
-0.017 |
| 530 |
248036_at |
AT5G55920
|
OLI2, OLIGOCELLULA 2 |
-0.217 |
0.120 |
| 531 |
257755_at |
AT3G18760
|
[Translation elongation factor EF1B/ribosomal protein S6 family protein] |
-0.217 |
0.041 |
| 532 |
251722_at |
AT3G56200
|
[Transmembrane amino acid transporter family protein] |
-0.216 |
0.016 |
| 533 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.216 |
-0.027 |
| 534 |
252358_at |
AT3G48425
|
[DNAse I-like superfamily protein] |
-0.216 |
0.013 |
| 535 |
258155_at |
AT3G18130
|
RACK1C, receptor for activated C kinase 1C, RACK1C_AT, receptor for activated C kinase 1C |
-0.216 |
0.074 |
| 536 |
256962_at |
AT3G13560
|
[O-Glycosyl hydrolases family 17 protein] |
-0.215 |
-0.001 |
| 537 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
-0.215 |
0.026 |
| 538 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
-0.214 |
0.106 |
| 539 |
265464_at |
AT2G37080
|
RIP2, ROP interactive partner 2 |
-0.214 |
0.009 |
| 540 |
245075_at |
AT2G23180
|
CYP96A1, cytochrome P450, family 96, subfamily A, polypeptide 1 |
-0.214 |
0.242 |
| 541 |
266581_at |
AT2G46140
|
[Late embryogenesis abundant protein] |
-0.213 |
-0.072 |
| 542 |
246249_at |
AT4G36680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.213 |
0.039 |
| 543 |
267586_at |
AT2G41950
|
unknown |
-0.212 |
0.036 |
| 544 |
267146_at |
AT2G38160
|
unknown |
-0.212 |
0.080 |
| 545 |
246157_at |
AT5G20080
|
[FAD/NAD(P)-binding oxidoreductase] |
-0.211 |
0.020 |
| 546 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.211 |
-0.011 |
| 547 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
-0.211 |
0.001 |
| 548 |
258521_at |
AT3G06680
|
[Ribosomal L29e protein family] |
-0.210 |
0.041 |
| 549 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
-0.210 |
-0.073 |
| 550 |
262313_at |
AT1G70900
|
unknown |
-0.210 |
-0.020 |
| 551 |
255348_at |
AT4G03820
|
unknown |
-0.210 |
0.078 |
| 552 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-0.210 |
-0.228 |
| 553 |
266297_at |
AT2G29570
|
ATPCNA2, A. THALIANA PROLIFERATING CELL NUCLEAR ANTIGEN 2, PCNA2, proliferating cell nuclear antigen 2 |
-0.209 |
0.040 |
| 554 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.209 |
0.106 |
| 555 |
263640_at |
AT2G25270
|
unknown |
-0.209 |
0.071 |
| 556 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
-0.209 |
0.069 |
| 557 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.208 |
0.046 |
| 558 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
-0.207 |
0.011 |
| 559 |
248854_at |
AT5G46580
|
[pentatricopeptide (PPR) repeat-containing protein] |
-0.206 |
-0.018 |
| 560 |
260392_at |
AT1G74030
|
ENO1, enolase 1 |
-0.206 |
0.052 |
| 561 |
259806_at |
AT1G47900
|
unknown |
-0.205 |
0.009 |
| 562 |
253445_at |
AT4G32605
|
[Mitochondrial glycoprotein family protein] |
-0.205 |
0.025 |
| 563 |
251142_at |
AT5G01015
|
unknown |
-0.204 |
-0.204 |
| 564 |
263136_at |
AT1G78580
|
ATTPS1, trehalose-6-phosphate synthase, TPS1, trehalose-6-phosphate synthase, TPS1, TREHALOSE-6-PHOSPHATE SYNTHASE 1 |
-0.204 |
-0.018 |
| 565 |
256150_at |
AT1G55120
|
AtcwINV3, 6-fructan exohydrolase, ATFRUCT5, beta-fructofuranosidase 5, FRUCT5, beta-fructofuranosidase 5 |
-0.203 |
0.059 |
| 566 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.202 |
-0.082 |
| 567 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
-0.202 |
-0.056 |
| 568 |
250994_at |
AT5G02490
|
AtHsp70-2, Hsp70-2 |
-0.202 |
0.037 |
| 569 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
-0.201 |
-0.001 |
| 570 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
-0.201 |
0.007 |
| 571 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
-0.201 |
0.046 |
| 572 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
-0.199 |
0.033 |
| 573 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.199 |
0.035 |
| 574 |
245341_at |
AT4G16447
|
unknown |
-0.198 |
-0.079 |
| 575 |
254109_at |
AT4G25240
|
SKS1, SKU5 similar 1 |
-0.198 |
0.032 |
| 576 |
261363_at |
AT1G41830
|
SKS6, SKU5 SIMILAR 6, SKS6, SKU5-similar 6 |
-0.198 |
-0.050 |
| 577 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.198 |
-0.024 |
| 578 |
262402_at |
AT1G49410
|
TOM6, translocase of the outer mitochondrial membrane 6 |
-0.198 |
0.016 |
| 579 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.198 |
-0.046 |
| 580 |
257005_at |
AT3G14190
|
CMR1, COPPER MODIFIED RESISTANCE 1 |
-0.197 |
0.066 |
| 581 |
259930_at |
AT1G34355
|
ATPS1, PARALLEL SPINDLE 1, PS1, PARALLEL SPINDLE 1 |
-0.197 |
0.073 |
| 582 |
250261_at |
AT5G13400
|
[Major facilitator superfamily protein] |
-0.196 |
-0.034 |
| 583 |
260714_at |
AT1G14980
|
CPN10, chaperonin 10 |
-0.196 |
0.022 |
| 584 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.196 |
-0.011 |
| 585 |
262533_at |
AT1G17090
|
unknown |
-0.196 |
-0.107 |
| 586 |
262181_at |
AT1G78060
|
[Glycosyl hydrolase family protein] |
-0.196 |
0.055 |
| 587 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.195 |
-0.173 |
| 588 |
252410_at |
AT3G47450
|
ATNOA1, ATNOS1, NOA1, NO ASSOCIATED 1, NOS1, NITRIC OXIDE SYNTHASE 1, RIF1, RESISTANT TO INHIBITION WITH FOSMIDOMYCIN 1 |
-0.194 |
0.001 |
| 589 |
247192_at |
AT5G65360
|
H3.1, histone 3.1 |
-0.194 |
0.005 |
| 590 |
253353_at |
AT4G33730
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.194 |
-0.439 |
| 591 |
261813_at |
AT1G08280
|
[Glycosyltransferase family 29 (sialyltransferase) family protein] |
-0.194 |
0.031 |
| 592 |
257201_at |
AT3G23740
|
unknown |
-0.193 |
-0.115 |
| 593 |
260100_at |
AT1G73177
|
APC13, anaphase-promoting complex 13, BNS, BONSAI |
-0.193 |
-0.012 |
| 594 |
266237_at |
AT2G29540
|
ATRPAC14, ATRPC14, RNApolymerase 14 kDa subunit, RPC14, RNApolymerase 14 kDa subunit |
-0.193 |
0.034 |
| 595 |
257822_at |
AT3G25230
|
ATFKBP62, FKBP62, FK506 BINDING PROTEIN 62, ROF1, rotamase FKBP 1 |
-0.193 |
0.020 |
| 596 |
253107_at |
AT4G35880
|
[Eukaryotic aspartyl protease family protein] |
-0.192 |
0.017 |
| 597 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.192 |
-0.658 |
| 598 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
-0.191 |
-0.009 |
| 599 |
253372_at |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
-0.191 |
-0.031 |
| 600 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.191 |
0.032 |