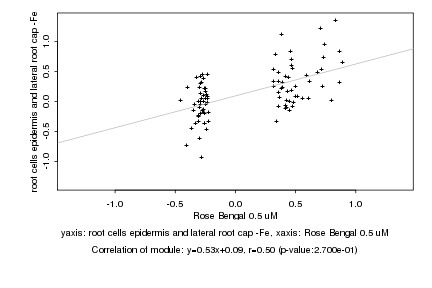
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Rose Bengal 0.5 uM |
Log2 signal ratio
root cells epidermis and lateral root cap -Fe |
| 1 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.887 |
0.659 |
| 2 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.864 |
0.848 |
| 3 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.860 |
0.330 |
| 4 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.826 |
1.368 |
| 5 |
245755_at |
AT1G35210
|
unknown |
0.798 |
0.025 |
| 6 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.739 |
0.959 |
| 7 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.729 |
0.735 |
| 8 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.722 |
0.264 |
| 9 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.711 |
0.548 |
| 10 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.702 |
1.234 |
| 11 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.680 |
0.486 |
| 12 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.608 |
0.344 |
| 13 |
263182_at |
AT1G05575
|
unknown |
0.605 |
0.063 |
| 14 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.586 |
0.444 |
| 15 |
257482_x_at |
AT1G27820
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.549 |
0.060 |
| 16 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.510 |
0.097 |
| 17 |
265327_at |
AT2G18210
|
unknown |
0.496 |
0.089 |
| 18 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.493 |
0.261 |
| 19 |
252567_at |
AT3G46070
|
[C2H2-type zinc finger family protein] |
0.481 |
-0.004 |
| 20 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.473 |
-0.081 |
| 21 |
265276_at |
AT2G28400
|
unknown |
0.469 |
0.565 |
| 22 |
264202_at |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
0.465 |
0.714 |
| 23 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.462 |
0.607 |
| 24 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
0.459 |
0.186 |
| 25 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.453 |
0.837 |
| 26 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.446 |
-0.149 |
| 27 |
256518_at |
AT1G66080
|
unknown |
0.443 |
0.014 |
| 28 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.440 |
0.416 |
| 29 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
0.432 |
0.179 |
| 30 |
249485_at |
AT5G39020
|
[Malectin/receptor-like protein kinase family protein] |
0.424 |
-0.084 |
| 31 |
252681_at |
AT3G44350
|
anac061, NAC domain containing protein 61, NAC061, NAC domain containing protein 61 |
0.418 |
0.023 |
| 32 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.414 |
-0.051 |
| 33 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.413 |
-0.115 |
| 34 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.409 |
0.426 |
| 35 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.387 |
0.239 |
| 36 |
262072_at |
AT1G59590
|
ZCF37 |
0.384 |
0.318 |
| 37 |
266092_at |
AT2G37880
|
unknown |
0.378 |
0.225 |
| 38 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.377 |
1.127 |
| 39 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.365 |
0.075 |
| 40 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.356 |
0.489 |
| 41 |
248286_at |
AT5G52870
|
MAKR5, MEMBRANE-ASSOCIATED KINASE REGULATOR 5 |
0.355 |
-0.074 |
| 42 |
264314_at |
AT1G70420
|
unknown |
0.355 |
0.338 |
| 43 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.352 |
0.164 |
| 44 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
0.338 |
-0.317 |
| 45 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.331 |
0.793 |
| 46 |
267065_at |
AT2G41080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.313 |
0.264 |
| 47 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.312 |
0.536 |
| 48 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
0.310 |
0.341 |
| 49 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
-0.458 |
0.033 |
| 50 |
250200_at |
AT5G14130
|
[Peroxidase superfamily protein] |
-0.412 |
-0.724 |
| 51 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
-0.402 |
0.236 |
| 52 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
-0.370 |
-0.437 |
| 53 |
246567_at |
AT5G14950
|
ATGMII, GMII, golgi alpha-mannosidase II |
-0.349 |
-0.140 |
| 54 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
-0.343 |
-0.049 |
| 55 |
266601_at |
AT2G46060
|
[transmembrane protein-related] |
-0.332 |
-0.362 |
| 56 |
252439_at |
AT3G47400
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.325 |
0.417 |
| 57 |
253925_at |
AT4G26690
|
GDPDL3, Glycerophosphodiester phosphodiesterase (GDPD) like 3, GPDL2, GLYCEROPHOSPHODIESTERASE-LIKE 2, MRH5, MUTANT ROOT HAIR 5, SHV3, SHAVEN 3 |
-0.314 |
-0.317 |
| 58 |
252128_at |
AT3G50870
|
GATA18, GATA TRANSCRIPTION FACTOR 18, HAN, HANABA TANARU, MNP, MONOPOLE |
-0.311 |
-0.228 |
| 59 |
261675_at |
AT1G18290
|
unknown |
-0.310 |
-0.246 |
| 60 |
256737_at |
AT3G29420
|
[geranylgeranyl pyrophosphate synthase-related / GGPP synthetase-related / farnesyltranstransferase-related] |
-0.308 |
0.015 |
| 61 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
-0.303 |
0.238 |
| 62 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.299 |
-0.615 |
| 63 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-0.299 |
-0.088 |
| 64 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-0.298 |
-0.184 |
| 65 |
258722_at |
AT3G09590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.297 |
0.007 |
| 66 |
263688_at |
AT1G26920
|
unknown |
-0.295 |
0.145 |
| 67 |
264389_at |
AT1G11960
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.293 |
0.422 |
| 68 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
-0.293 |
0.304 |
| 69 |
266022_at |
AT2G05920
|
[Subtilase family protein] |
-0.291 |
-0.038 |
| 70 |
253875_at |
AT4G27520
|
AtENODL2, ENODL2, early nodulin-like protein 2 |
-0.290 |
0.064 |
| 71 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
-0.289 |
0.325 |
| 72 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.287 |
-0.924 |
| 73 |
245436_at |
AT4G16620
|
UMAMIT8, Usually multiple acids move in and out Transporters 8 |
-0.281 |
0.463 |
| 74 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.276 |
-0.148 |
| 75 |
256908_at |
AT3G24040
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.272 |
0.005 |
| 76 |
262846_at |
AT1G14670
|
[Endomembrane protein 70 protein family] |
-0.270 |
0.102 |
| 77 |
265994_at |
AT2G24170
|
[Endomembrane protein 70 protein family] |
-0.270 |
0.387 |
| 78 |
263293_x_at |
AT2G10880
|
[hypothetical protein, similar to hypothetical protein GB:AAC26673] |
-0.269 |
-0.176 |
| 79 |
264136_at |
AT1G78980
|
SRF5, STRUBBELIG-receptor family 5 |
-0.267 |
-0.129 |
| 80 |
254340_at |
AT4G22120
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.265 |
-0.351 |
| 81 |
248572_at |
AT5G49800
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.262 |
-0.187 |
| 82 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.261 |
0.220 |
| 83 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
-0.260 |
-0.359 |
| 84 |
255189_at |
AT4G07350
|
unknown |
-0.257 |
-0.039 |
| 85 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.253 |
0.222 |
| 86 |
262768_at |
AT1G12990
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.252 |
0.181 |
| 87 |
250143_at |
AT5G14670
|
ARFA1B, ADP-ribosylation factor A1B, ATARFA1B, ADP-ribosylation factor A1B |
-0.250 |
0.055 |
| 88 |
264001_at |
AT2G22420
|
[Peroxidase superfamily protein] |
-0.244 |
-0.462 |
| 89 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
-0.241 |
0.132 |
| 90 |
262504_at |
AT1G21750
|
ATPDI5, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 5, ATPDIL1-1, PDI-like 1-1, PDI5, PROTEIN DISULFIDE ISOMERASE 5, PDIL1-1, PDI-like 1-1 |
-0.240 |
-0.011 |
| 91 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.235 |
0.452 |
| 92 |
255332_at |
AT4G04340
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.234 |
0.057 |
| 93 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
-0.234 |
0.085 |
| 94 |
254109_at |
AT4G25240
|
SKS1, SKU5 similar 1 |
-0.231 |
-0.180 |
| 95 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
-0.227 |
-0.323 |