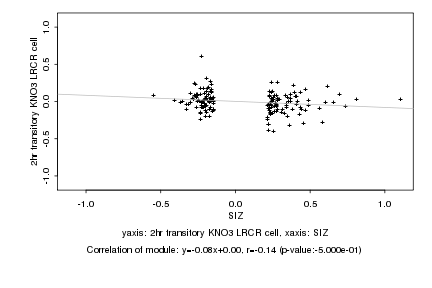
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
SIZ |
Log2 signal ratio
2hr transitory KNO3 LRCR cell |
| 1 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
1.102 |
0.038 |
| 2 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.804 |
0.033 |
| 3 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.730 |
-0.058 |
| 4 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.696 |
0.106 |
| 5 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.655 |
-0.001 |
| 6 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.613 |
0.202 |
| 7 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
0.602 |
-0.002 |
| 8 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.580 |
-0.275 |
| 9 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
0.557 |
-0.093 |
| 10 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.487 |
0.015 |
| 11 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.487 |
-0.043 |
| 12 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
0.466 |
0.166 |
| 13 |
259864_at |
AT1G72800
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.465 |
-0.112 |
| 14 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.452 |
-0.287 |
| 15 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
0.437 |
-0.096 |
| 16 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.432 |
0.131 |
| 17 |
258178_at |
AT3G21680
|
unknown |
0.430 |
-0.070 |
| 18 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.426 |
-0.166 |
| 19 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.413 |
0.006 |
| 20 |
260472_at |
AT1G10990
|
unknown |
0.406 |
-0.039 |
| 21 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
0.403 |
0.079 |
| 22 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.401 |
0.068 |
| 23 |
265327_at |
AT2G18210
|
unknown |
0.389 |
0.124 |
| 24 |
261221_at |
AT1G19960
|
unknown |
0.388 |
0.225 |
| 25 |
249394_at |
AT5G40180
|
unknown |
0.377 |
-0.100 |
| 26 |
260092_at |
AT1G73280
|
scpl3, serine carboxypeptidase-like 3 |
0.365 |
0.008 |
| 27 |
248822_at |
AT5G47000
|
[Peroxidase superfamily protein] |
0.363 |
0.103 |
| 28 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.362 |
0.042 |
| 29 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.361 |
-0.320 |
| 30 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.355 |
0.007 |
| 31 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.348 |
-0.199 |
| 32 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.345 |
0.055 |
| 33 |
250474_at |
AT5G10230
|
ANN7, ANNEXIN 7, ANNAT7, annexin 7 |
0.339 |
-0.025 |
| 34 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.339 |
-0.069 |
| 35 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.338 |
-0.041 |
| 36 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.329 |
0.084 |
| 37 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
0.327 |
-0.151 |
| 38 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
0.313 |
-0.103 |
| 39 |
264987_at |
AT1G27030
|
unknown |
0.306 |
-0.143 |
| 40 |
246072_at |
AT5G20240
|
PI, PISTILLATA |
0.291 |
0.034 |
| 41 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.285 |
0.028 |
| 42 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.281 |
0.017 |
| 43 |
264912_at |
AT1G60750
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.277 |
-0.100 |
| 44 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.276 |
0.257 |
| 45 |
249485_at |
AT5G39020
|
[Malectin/receptor-like protein kinase family protein] |
0.276 |
0.043 |
| 46 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
0.273 |
-0.046 |
| 47 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.272 |
0.092 |
| 48 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.272 |
-0.056 |
| 49 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.269 |
-0.029 |
| 50 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.269 |
-0.125 |
| 51 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
0.268 |
-0.014 |
| 52 |
259952_at |
AT1G71400
|
AtRLP12, receptor like protein 12, RLP12, receptor like protein 12 |
0.268 |
-0.024 |
| 53 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.261 |
0.083 |
| 54 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.257 |
-0.126 |
| 55 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
0.256 |
-0.057 |
| 56 |
253112_at |
AT4G35970
|
APX5, ascorbate peroxidase 5 |
0.253 |
0.062 |
| 57 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.252 |
-0.391 |
| 58 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.249 |
0.035 |
| 59 |
255945_at |
AT5G28610
|
unknown |
0.247 |
0.141 |
| 60 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
0.246 |
0.001 |
| 61 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
0.244 |
0.040 |
| 62 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
0.242 |
-0.148 |
| 63 |
250610_at |
AT5G07550
|
ATGRP19, GRP19, glycine-rich protein 19 |
0.241 |
0.131 |
| 64 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
0.240 |
0.268 |
| 65 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.238 |
-0.048 |
| 66 |
265433_at |
AT2G20950
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.238 |
-0.076 |
| 67 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.237 |
0.052 |
| 68 |
247742_at |
AT5G58980
|
[Neutral/alkaline non-lysosomal ceramidase] |
0.237 |
0.007 |
| 69 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
0.234 |
-0.150 |
| 70 |
256617_at |
AT3G22240
|
unknown |
0.234 |
-0.035 |
| 71 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.229 |
-0.038 |
| 72 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
0.229 |
-0.001 |
| 73 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
0.229 |
-0.164 |
| 74 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.228 |
-0.146 |
| 75 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
0.228 |
-0.137 |
| 76 |
262913_at |
AT1G59960
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.227 |
0.140 |
| 77 |
250472_at |
AT5G10210
|
unknown |
0.227 |
0.089 |
| 78 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.226 |
-0.111 |
| 79 |
266581_at |
AT2G46140
|
[Late embryogenesis abundant protein] |
0.225 |
-0.091 |
| 80 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.223 |
0.073 |
| 81 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.222 |
-0.048 |
| 82 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.221 |
-0.093 |
| 83 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.220 |
-0.379 |
| 84 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.219 |
-0.296 |
| 85 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.217 |
-0.306 |
| 86 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.217 |
-0.037 |
| 87 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
0.213 |
-0.049 |
| 88 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.209 |
-0.204 |
| 89 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
0.208 |
-0.234 |
| 90 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.549 |
0.094 |
| 91 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.410 |
0.014 |
| 92 |
262301_at |
AT1G70880
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.371 |
-0.008 |
| 93 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.358 |
0.009 |
| 94 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
-0.334 |
-0.037 |
| 95 |
248394_at |
AT5G52070
|
[Agenet domain-containing protein] |
-0.334 |
-0.099 |
| 96 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.320 |
-0.027 |
| 97 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.304 |
-0.012 |
| 98 |
264958_at |
AT1G76960
|
unknown |
-0.303 |
0.119 |
| 99 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.293 |
0.045 |
| 100 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
-0.281 |
0.030 |
| 101 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.280 |
0.250 |
| 102 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.274 |
0.088 |
| 103 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
-0.271 |
0.079 |
| 104 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.270 |
0.240 |
| 105 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.266 |
-0.079 |
| 106 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
-0.262 |
0.063 |
| 107 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.261 |
0.107 |
| 108 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.258 |
0.002 |
| 109 |
245420_at |
AT4G17410
|
[DWNN domain, a CCHC-type zinc finger] |
-0.257 |
0.102 |
| 110 |
259281_at |
AT3G01140
|
AtMYB106, myb domain protein 106, MYB106, myb domain protein 106, NOK, NOECK |
-0.254 |
0.085 |
| 111 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.251 |
0.020 |
| 112 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
-0.244 |
0.005 |
| 113 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
-0.239 |
-0.135 |
| 114 |
267204_at |
AT2G31050
|
[Cupredoxin superfamily protein] |
-0.239 |
-0.155 |
| 115 |
247126_at |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-0.237 |
0.095 |
| 116 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
-0.237 |
-0.230 |
| 117 |
260771_at |
AT1G49160
|
WNK7 |
-0.234 |
0.181 |
| 118 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
-0.233 |
-0.050 |
| 119 |
251102_at |
AT5G01690
|
ATCHX27, ARABIDOPSIS THALIANA CATION/HYDROGEN EXCHANGER 27, CHX27, cation/H+ exchanger 27 |
-0.232 |
-0.070 |
| 120 |
257451_at |
AT1G05690
|
BT3, BTB and TAZ domain protein 3 |
-0.231 |
-0.001 |
| 121 |
260717_at |
AT1G48120
|
[hydrolases] |
-0.229 |
-0.010 |
| 122 |
253084_at |
AT4G36260
|
SRS2, SHI RELATED SEQUENCE 2, STY2, STYLISH 2 |
-0.228 |
0.613 |
| 123 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.226 |
-0.077 |
| 124 |
262388_at |
AT1G49320
|
unknown |
-0.225 |
-0.001 |
| 125 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.218 |
-0.076 |
| 126 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.214 |
0.177 |
| 127 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.214 |
0.020 |
| 128 |
247553_at |
AT5G60910
|
AGL8, AGAMOUS-like 8, FUL, FRUITFULL |
-0.213 |
0.035 |
| 129 |
249069_at |
AT5G44010
|
unknown |
-0.213 |
0.109 |
| 130 |
260156_at |
AT1G52880
|
ANAC018, Arabidopsis NAC domain containing protein 18, ATNAM, NAM, NO APICAL MERISTEM, NARS2, NAC-REGULATED SEED MORPHOLOGY 2 |
-0.212 |
-0.022 |
| 131 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.210 |
-0.022 |
| 132 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-0.207 |
0.044 |
| 133 |
244973_at |
ATCG00690
|
PSBT, photosystem II reaction center protein T, PSBTC |
-0.206 |
-0.059 |
| 134 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
-0.205 |
-0.044 |
| 135 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.204 |
-0.115 |
| 136 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
-0.204 |
-0.041 |
| 137 |
249094_at |
AT5G43890
|
SUPER1, SUPPRESSOR OF ER 1, YUC5, YUCCA5 |
-0.202 |
-0.189 |
| 138 |
267547_at |
AT2G32670
|
ATVAMP725, vesicle-associated membrane protein 725, VAMP725, vesicle-associated membrane protein 725 |
-0.199 |
0.143 |
| 139 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.198 |
-0.047 |
| 140 |
261336_at |
AT1G44790
|
[ChaC-like family protein] |
-0.197 |
0.312 |
| 141 |
261074_at |
AT1G07290
|
GONST2, golgi nucleotide sugar transporter 2 |
-0.195 |
-0.139 |
| 142 |
256021_at |
AT1G58270
|
ZW9 |
-0.194 |
0.053 |
| 143 |
259350_at |
AT3G05140
|
RBK2, ROP binding protein kinases 2 |
-0.194 |
0.080 |
| 144 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.193 |
0.187 |
| 145 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.187 |
0.026 |
| 146 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
-0.186 |
0.198 |
| 147 |
262214_at |
AT1G74820
|
[RmlC-like cupins superfamily protein] |
-0.182 |
0.095 |
| 148 |
252304_at |
AT3G49230
|
unknown |
-0.179 |
-0.100 |
| 149 |
250483_at |
AT5G10300
|
AtHNL, ATMES5, ARABIDOPSIS THALIANA METHYL ESTERASE 5, HNL, HYDROXYNITRILE LYASE, MES5, methyl esterase 5 |
-0.176 |
0.010 |
| 150 |
267025_at |
AT2G38350
|
unknown |
-0.176 |
-0.191 |
| 151 |
254745_at |
AT4G13460
|
SDG22, SETDOMAIN GROUP 22, SET22, SUVH9, SU(VAR)3-9 homolog 9 |
-0.175 |
-0.001 |
| 152 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
-0.174 |
-0.101 |
| 153 |
251302_at |
AT3G61970
|
NGA2, NGATHA2 |
-0.174 |
0.021 |
| 154 |
262067_at |
AT1G80060
|
[Ubiquitin-like superfamily protein] |
-0.173 |
0.059 |
| 155 |
249856_at |
AT5G22980
|
scpl47, serine carboxypeptidase-like 47 |
-0.172 |
0.042 |
| 156 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
-0.172 |
0.024 |
| 157 |
267535_at |
AT2G41940
|
ZFP8, zinc finger protein 8 |
-0.171 |
0.139 |
| 158 |
263754_at |
AT2G21510
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.170 |
0.005 |
| 159 |
254008_at |
AT4G26380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.170 |
0.270 |
| 160 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.168 |
0.021 |
| 161 |
246568_at |
AT5G14960
|
DEL2, DP-E2F-like 2, E2FD, E2L1 |
-0.167 |
-0.079 |
| 162 |
246757_at |
AT5G27940
|
WPP3, WPP domain protein 3 |
-0.164 |
0.165 |
| 163 |
246807_at |
AT5G27100
|
ATGLR2.1, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 2.1, GLR2.1, glutamate receptor 2.1 |
-0.164 |
0.022 |
| 164 |
255909_at |
AT1G18040
|
AT;CDCKD;3, CYCLIN-DEPENDENT KINASE D1;3, CAK2AT, CDKD1;3, cyclin-dependent kinase D1;3 |
-0.163 |
0.141 |
| 165 |
254450_at |
AT4G21080
|
AtDOF4.5, DOF4.5, DNA-binding with One Finger 4.5 |
-0.163 |
0.037 |
| 166 |
246118_at |
AT5G20340
|
BG5, beta-1,3-glucanase 5 |
-0.163 |
0.127 |
| 167 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
-0.162 |
0.039 |
| 168 |
264843_at |
AT1G03400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.161 |
0.231 |
| 169 |
260583_x_at |
AT2G47220
|
unknown |
-0.156 |
0.054 |
| 170 |
245901_at |
AT5G11060
|
KNAT4, KNOTTED1-like homeobox gene 4 |
-0.156 |
-0.123 |
| 171 |
260843_at |
AT1G29060
|
[Target SNARE coiled-coil domain protein] |
-0.153 |
-0.025 |
| 172 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.152 |
-0.101 |
| 173 |
265234_at |
AT2G07721
|
unknown |
-0.152 |
0.009 |
| 174 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.151 |
-0.113 |
| 175 |
252178_at |
AT3G50750
|
BEH1, BES1/BZR1 homolog 1 |
-0.150 |
0.063 |
| 176 |
266853_at |
AT2G26790
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.149 |
0.021 |
| 177 |
264208_at |
AT1G22760
|
PAB3, poly(A) binding protein 3 |
-0.149 |
-0.102 |