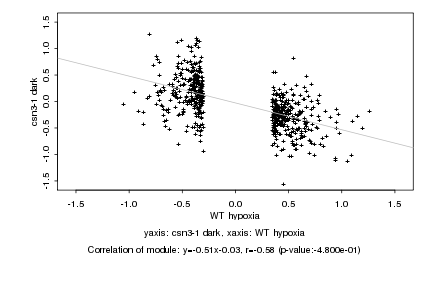
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT hypoxia |
Log2 signal ratio
csn3-1 dark |
| 1 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
1.252 |
-0.186 |
| 2 |
265387_at |
AT2G20670
|
unknown |
1.194 |
-0.504 |
| 3 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
1.141 |
-0.276 |
| 4 |
245076_at |
AT2G23170
|
GH3.3 |
1.094 |
-0.372 |
| 5 |
247474_at |
AT5G62280
|
unknown |
1.091 |
-1.007 |
| 6 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
1.053 |
-1.115 |
| 7 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.975 |
-0.590 |
| 8 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.964 |
-0.239 |
| 9 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.946 |
-0.144 |
| 10 |
250327_at |
AT5G12050
|
unknown |
0.946 |
-0.393 |
| 11 |
252882_at |
AT4G39675
|
unknown |
0.944 |
-0.505 |
| 12 |
251012_at |
AT5G02580
|
unknown |
0.941 |
-1.071 |
| 13 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
0.939 |
-1.104 |
| 14 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
0.886 |
-0.285 |
| 15 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.849 |
-0.647 |
| 16 |
261567_at |
AT1G33055
|
unknown |
0.843 |
-0.179 |
| 17 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.824 |
-0.846 |
| 18 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.814 |
-0.687 |
| 19 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.800 |
-0.793 |
| 20 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
0.787 |
-0.357 |
| 21 |
264323_at |
AT1G04180
|
YUC9, YUCCA 9 |
0.783 |
0.128 |
| 22 |
265276_at |
AT2G28400
|
unknown |
0.773 |
-0.272 |
| 23 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.772 |
-0.035 |
| 24 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.768 |
0.031 |
| 25 |
260668_at |
AT1G19530
|
unknown |
0.765 |
-0.491 |
| 26 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.758 |
-0.476 |
| 27 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.742 |
-1.017 |
| 28 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.741 |
-0.806 |
| 29 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.728 |
-0.102 |
| 30 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.724 |
-0.433 |
| 31 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.714 |
-0.791 |
| 32 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.709 |
-0.138 |
| 33 |
259015_at |
AT3G07350
|
unknown |
0.709 |
-0.257 |
| 34 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.708 |
-0.538 |
| 35 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.707 |
0.324 |
| 36 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.707 |
0.017 |
| 37 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.705 |
-0.748 |
| 38 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.696 |
-0.976 |
| 39 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
0.692 |
-0.725 |
| 40 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.685 |
-0.515 |
| 41 |
250152_at |
AT5G15120
|
unknown |
0.678 |
0.105 |
| 42 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.672 |
-0.394 |
| 43 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
0.668 |
-0.471 |
| 44 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.667 |
-0.099 |
| 45 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.665 |
0.483 |
| 46 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.665 |
0.198 |
| 47 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.663 |
-0.371 |
| 48 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.663 |
-0.105 |
| 49 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.656 |
-0.431 |
| 50 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.655 |
-0.412 |
| 51 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.655 |
-0.555 |
| 52 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.646 |
0.279 |
| 53 |
264238_at |
AT1G54740
|
unknown |
0.646 |
-0.310 |
| 54 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.645 |
0.042 |
| 55 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.645 |
-0.649 |
| 56 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.643 |
-0.506 |
| 57 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
0.641 |
-0.366 |
| 58 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.634 |
0.051 |
| 59 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.628 |
-0.182 |
| 60 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.625 |
-0.661 |
| 61 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.617 |
-0.553 |
| 62 |
260744_at |
AT1G15010
|
unknown |
0.617 |
-0.586 |
| 63 |
248509_at |
AT5G50335
|
unknown |
0.616 |
-0.815 |
| 64 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.615 |
-0.623 |
| 65 |
251246_at |
AT3G62100
|
IAA30, indole-3-acetic acid inducible 30 |
0.609 |
-0.204 |
| 66 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
0.609 |
-0.190 |
| 67 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.608 |
-0.685 |
| 68 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.608 |
0.130 |
| 69 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.603 |
-0.216 |
| 70 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
0.601 |
-0.504 |
| 71 |
258383_at |
AT3G15440
|
unknown |
0.600 |
-0.188 |
| 72 |
258262_at |
AT3G15770
|
unknown |
0.596 |
-0.233 |
| 73 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.592 |
-0.501 |
| 74 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
0.591 |
-0.005 |
| 75 |
261456_at |
AT1G21050
|
unknown |
0.590 |
-0.345 |
| 76 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
0.590 |
-0.355 |
| 77 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.587 |
-0.253 |
| 78 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.587 |
-0.354 |
| 79 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.585 |
-0.055 |
| 80 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.583 |
-0.250 |
| 81 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.578 |
-0.329 |
| 82 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.576 |
-0.542 |
| 83 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.574 |
-0.901 |
| 84 |
247878_at |
AT5G57760
|
unknown |
0.573 |
-0.802 |
| 85 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.573 |
-0.335 |
| 86 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.569 |
0.309 |
| 87 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.568 |
-0.512 |
| 88 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.566 |
-0.714 |
| 89 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.563 |
-0.027 |
| 90 |
261451_at |
AT1G21060
|
unknown |
0.563 |
-0.237 |
| 91 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.562 |
-0.896 |
| 92 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.561 |
-0.594 |
| 93 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
0.560 |
-0.397 |
| 94 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.555 |
-0.309 |
| 95 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
0.547 |
-0.817 |
| 96 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.546 |
-0.365 |
| 97 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
0.543 |
-0.561 |
| 98 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.543 |
0.824 |
| 99 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
0.542 |
0.128 |
| 100 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.541 |
-0.815 |
| 101 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.539 |
-0.338 |
| 102 |
261387_at |
AT1G05410
|
unknown |
0.538 |
0.134 |
| 103 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.536 |
-0.495 |
| 104 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
0.535 |
0.021 |
| 105 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.533 |
-0.742 |
| 106 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.532 |
-0.274 |
| 107 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
0.531 |
-0.792 |
| 108 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.529 |
-0.120 |
| 109 |
254193_at |
AT4G23870
|
unknown |
0.526 |
-1.023 |
| 110 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.523 |
-0.112 |
| 111 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.522 |
-0.643 |
| 112 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.519 |
-0.583 |
| 113 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.518 |
0.228 |
| 114 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.516 |
-0.650 |
| 115 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.515 |
-0.226 |
| 116 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.513 |
-0.076 |
| 117 |
260760_at |
AT1G49170
|
unknown |
0.512 |
-0.168 |
| 118 |
253155_at |
AT4G35720
|
unknown |
0.504 |
-1.024 |
| 119 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.502 |
-0.608 |
| 120 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.501 |
-0.319 |
| 121 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.500 |
-0.630 |
| 122 |
246929_at |
AT5G25210
|
unknown |
0.499 |
-0.349 |
| 123 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.498 |
0.027 |
| 124 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
0.498 |
-0.035 |
| 125 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.495 |
-0.173 |
| 126 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.494 |
-0.514 |
| 127 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.494 |
-0.170 |
| 128 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.494 |
-0.245 |
| 129 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.492 |
-0.141 |
| 130 |
263799_at |
AT2G24550
|
unknown |
0.489 |
-0.353 |
| 131 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
0.486 |
-0.051 |
| 132 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.484 |
-0.317 |
| 133 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.484 |
-0.155 |
| 134 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.484 |
-0.321 |
| 135 |
266259_at |
AT2G27830
|
unknown |
0.482 |
-0.470 |
| 136 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.479 |
-0.233 |
| 137 |
248801_at |
AT5G47370
|
HAT2 |
0.478 |
-0.618 |
| 138 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.478 |
-0.202 |
| 139 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.475 |
0.184 |
| 140 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.471 |
0.034 |
| 141 |
264529_at |
AT1G30820
|
[CTP synthase family protein] |
0.470 |
-0.096 |
| 142 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.469 |
-0.281 |
| 143 |
247597_at |
AT5G60860
|
AtRABA1f, RAB GTPase homolog A1F, RABA1f, RAB GTPase homolog A1F |
0.469 |
-0.220 |
| 144 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
0.466 |
-0.542 |
| 145 |
263118_at |
AT1G03090
|
MCCA |
0.465 |
-0.361 |
| 146 |
264447_at |
AT1G27300
|
unknown |
0.465 |
0.000 |
| 147 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
0.464 |
-0.003 |
| 148 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
0.462 |
-0.223 |
| 149 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.458 |
0.339 |
| 150 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.457 |
-0.420 |
| 151 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.457 |
-0.041 |
| 152 |
254178_at |
AT4G23880
|
unknown |
0.456 |
-0.303 |
| 153 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.456 |
-0.169 |
| 154 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.455 |
-0.079 |
| 155 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
0.455 |
-0.349 |
| 156 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.452 |
-0.753 |
| 157 |
247903_at |
AT5G57340
|
unknown |
0.452 |
-0.080 |
| 158 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
0.449 |
0.141 |
| 159 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.448 |
-0.335 |
| 160 |
264836_at |
AT1G03610
|
unknown |
0.447 |
-0.309 |
| 161 |
246215_at |
AT4G37180
|
[Homeodomain-like superfamily protein] |
0.446 |
0.089 |
| 162 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
0.446 |
-0.462 |
| 163 |
248279_at |
AT5G52910
|
ATIM, TIMELESS |
0.446 |
0.127 |
| 164 |
247754_at |
AT5G59080
|
unknown |
0.446 |
-0.903 |
| 165 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
0.445 |
-1.549 |
| 166 |
267077_at |
AT2G40970
|
MYBC1 |
0.444 |
-0.280 |
| 167 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
0.444 |
-0.261 |
| 168 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.443 |
-0.105 |
| 169 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
0.441 |
-0.444 |
| 170 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
0.440 |
-0.212 |
| 171 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.440 |
-0.145 |
| 172 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.440 |
-0.116 |
| 173 |
256114_at |
AT1G16850
|
unknown |
0.437 |
0.079 |
| 174 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.436 |
-0.357 |
| 175 |
247835_at |
AT5G57910
|
unknown |
0.435 |
-0.417 |
| 176 |
250937_at |
AT5G03230
|
unknown |
0.435 |
0.003 |
| 177 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.432 |
-0.256 |
| 178 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.430 |
-0.397 |
| 179 |
262801_at |
AT1G21010
|
unknown |
0.429 |
-0.183 |
| 180 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
0.429 |
-0.912 |
| 181 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.428 |
-0.387 |
| 182 |
264508_at |
AT1G09570
|
FHY2, FAR RED ELONGATED HYPOCOTYL 2, FRE1, FAR RED ELONGATED 1, HY8, ELONGATED HYPOCOTYL 8, PHYA, phytochrome A |
0.426 |
-0.294 |
| 183 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.425 |
-0.040 |
| 184 |
263914_at |
AT2G36400
|
AtGRF3, growth-regulating factor 3, GRF3, growth-regulating factor 3 |
0.425 |
-0.103 |
| 185 |
250464_at |
AT5G10040
|
unknown |
0.425 |
0.170 |
| 186 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.424 |
-0.443 |
| 187 |
247585_at |
AT5G60680
|
unknown |
0.422 |
-0.224 |
| 188 |
248585_at |
AT5G49640
|
unknown |
0.422 |
-0.235 |
| 189 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.422 |
-0.622 |
| 190 |
252424_at |
AT3G47610
|
[transcription regulators] |
0.421 |
0.085 |
| 191 |
249932_at |
AT5G22390
|
unknown |
0.419 |
-0.190 |
| 192 |
264580_at |
AT1G05340
|
unknown |
0.419 |
-0.441 |
| 193 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.418 |
0.035 |
| 194 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.414 |
-0.605 |
| 195 |
253404_at |
AT4G32840
|
PFK6, phosphofructokinase 6 |
0.414 |
0.117 |
| 196 |
255866_at |
AT2G30350
|
[Excinuclease ABC, C subunit, N-terminal] |
0.413 |
-0.277 |
| 197 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.413 |
-0.421 |
| 198 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.413 |
-0.369 |
| 199 |
264989_at |
AT1G27200
|
unknown |
0.412 |
-0.232 |
| 200 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.411 |
0.237 |
| 201 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
0.410 |
-0.103 |
| 202 |
248869_at |
AT5G46840
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.410 |
-0.297 |
| 203 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
0.409 |
0.128 |
| 204 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.408 |
-0.238 |
| 205 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.406 |
-0.167 |
| 206 |
247029_at |
AT5G67190
|
DEAR2, DREB and EAR motif protein 2 |
0.401 |
-0.345 |
| 207 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.401 |
-0.313 |
| 208 |
263931_at |
AT2G36220
|
unknown |
0.401 |
-0.222 |
| 209 |
246222_at |
AT4G36900
|
DEAR4, DREB AND EAR MOTIF PROTEIN 4, RAP2.10, related to AP2 10 |
0.400 |
-0.275 |
| 210 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
0.399 |
-0.085 |
| 211 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
0.397 |
-0.121 |
| 212 |
260310_at |
AT1G70590
|
[F-box family protein] |
0.397 |
-0.095 |
| 213 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.394 |
-0.446 |
| 214 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.394 |
-0.844 |
| 215 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.394 |
-0.448 |
| 216 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.393 |
-0.405 |
| 217 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
0.390 |
-0.255 |
| 218 |
257580_at |
AT3G06210
|
[ARM repeat superfamily protein] |
0.390 |
-0.235 |
| 219 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
0.390 |
-0.581 |
| 220 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
0.388 |
-0.065 |
| 221 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
0.388 |
0.146 |
| 222 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.387 |
-0.260 |
| 223 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
0.386 |
-0.342 |
| 224 |
247743_at |
AT5G59010
|
BSK5, brassinosteroid-signaling kinase 5 |
0.386 |
-0.309 |
| 225 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
0.385 |
-0.095 |
| 226 |
261580_at |
AT1G01110
|
IQD18, IQ-domain 18 |
0.385 |
-0.364 |
| 227 |
250603_at |
AT5G07820
|
[Plant calmodulin-binding protein-related] |
0.384 |
-0.191 |
| 228 |
261039_at |
AT1G17455
|
ELF4-L4, ELF4-like 4 |
0.384 |
-0.158 |
| 229 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.384 |
-0.709 |
| 230 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
0.384 |
-0.130 |
| 231 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
0.384 |
0.164 |
| 232 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.383 |
-0.179 |
| 233 |
261205_at |
AT1G12790
|
unknown |
0.383 |
-0.148 |
| 234 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
0.381 |
-0.065 |
| 235 |
254424_at |
AT4G21510
|
AtFBS2, FBS2, F-box stress induced 2 |
0.380 |
-0.333 |
| 236 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.380 |
-1.010 |
| 237 |
264379_at |
AT2G25200
|
unknown |
0.379 |
0.033 |
| 238 |
253802_at |
AT4G28180
|
unknown |
0.379 |
-0.220 |
| 239 |
254073_at |
AT4G25500
|
At-RS40, arginine/serine-rich splicing factor 40, AT-SRP40, ATRSP35, ARABIDOPSIS THALIANA ARGININE/SERINE-RICH SPLICING FACTOR 35, ATRSP40, RS40, arginine/serine-rich splicing factor 40, RSP35, arginine/serine-rich splicing factor 35 |
0.379 |
-0.119 |
| 240 |
249944_at |
AT5G22290
|
anac089, NAC domain containing protein 89, FSQ6, fructose-sensing quantitative trait locus 6, NAC089, NAC domain containing protein 89 |
0.379 |
-0.048 |
| 241 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.378 |
0.116 |
| 242 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.377 |
-0.530 |
| 243 |
265266_at |
AT2G42890
|
AML2, MEI2-like 2, ML2, MEI2-like 2 |
0.376 |
-0.386 |
| 244 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
0.376 |
-0.397 |
| 245 |
259230_at |
AT3G07780
|
OBE1, OBERON1 |
0.375 |
-0.231 |
| 246 |
263490_at |
AT2G42620
|
MAX2, MORE AXILLARY BRANCHES 2, ORE9, ORESARA 9, PPS, PLEIOTROPIC PHOTOSIGNALING |
0.375 |
-0.053 |
| 247 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.374 |
-0.246 |
| 248 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
0.371 |
-0.453 |
| 249 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.371 |
-0.621 |
| 250 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.370 |
-0.650 |
| 251 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.370 |
0.005 |
| 252 |
262952_at |
AT1G75770
|
unknown |
0.370 |
0.028 |
| 253 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
0.370 |
-0.529 |
| 254 |
250830_at |
AT5G04910
|
unknown |
0.369 |
0.024 |
| 255 |
251922_at |
AT3G54030
|
BSK6, brassinosteroid-signaling kinase 6 |
0.369 |
-0.035 |
| 256 |
252412_at |
AT3G47295
|
unknown |
0.369 |
-0.096 |
| 257 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
0.368 |
-0.118 |
| 258 |
250748_at |
AT5G05710
|
[Pleckstrin homology (PH) domain superfamily protein] |
0.368 |
-0.338 |
| 259 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.368 |
0.549 |
| 260 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
0.367 |
-0.470 |
| 261 |
263305_at |
AT2G01930
|
ATBPC1, BASIC PENTACYSTEINE1, BBR, BPC1, basic pentacysteine1 |
0.366 |
-0.297 |
| 262 |
262845_at |
AT1G14740
|
TTA1, TITANIA 1 |
0.366 |
-0.261 |
| 263 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.366 |
-0.280 |
| 264 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.365 |
-0.396 |
| 265 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.364 |
-0.192 |
| 266 |
267423_at |
AT2G35060
|
KUP11, K+ uptake permease 11 |
0.364 |
-0.220 |
| 267 |
247193_at |
AT5G65380
|
[MATE efflux family protein] |
0.364 |
-0.159 |
| 268 |
250975_at |
AT5G03050
|
unknown |
0.362 |
0.222 |
| 269 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.362 |
-0.506 |
| 270 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.361 |
-0.784 |
| 271 |
264264_at |
AT1G09250
|
AIF4, ATBS1 Interacting Factor 4 |
0.361 |
-0.307 |
| 272 |
257990_at |
AT3G19860
|
bHLH121, basic Helix-Loop-Helix 121 |
0.359 |
-0.225 |
| 273 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.359 |
0.000 |
| 274 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.359 |
-0.570 |
| 275 |
255854_at |
AT1G67050
|
unknown |
0.359 |
-0.415 |
| 276 |
245307_at |
AT4G16770
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.358 |
0.141 |
| 277 |
245353_at |
AT4G16000
|
unknown |
0.358 |
-0.566 |
| 278 |
259645_at |
AT1G69010
|
BIM2, BES1-interacting Myc-like protein 2 |
0.358 |
-0.240 |
| 279 |
264889_at |
AT1G23050
|
hydroxyproline-rich glycoprotein family protein |
0.357 |
-0.409 |
| 280 |
254210_at |
AT4G23450
|
AIRP1, ABA Insensitive RING Protein 1, AtAIRP1 |
0.356 |
-0.272 |
| 281 |
264774_at |
AT1G22890
|
unknown |
0.356 |
-0.261 |
| 282 |
260279_at |
AT1G80420
|
ATXRCC1, XRCC1, homolog of X-ray repair cross complementing 1 |
0.355 |
-0.200 |
| 283 |
259356_at |
AT3G05250
|
[RING/U-box superfamily protein] |
0.355 |
-0.144 |
| 284 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.354 |
-0.436 |
| 285 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.354 |
-0.609 |
| 286 |
257894_at |
AT3G17100
|
AIF3, ATBS1 Interacting Factor 3 |
0.352 |
-0.489 |
| 287 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.351 |
0.003 |
| 288 |
261866_at |
AT1G50420
|
SCL-3, SCARECROW-LIKE 3, SCL3, scarecrow-like 3 |
0.350 |
0.026 |
| 289 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.350 |
0.279 |
| 290 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.350 |
0.552 |
| 291 |
262204_at |
AT2G01100
|
unknown |
0.349 |
-0.196 |
| 292 |
253811_at |
AT4G28190
|
ULT, ULTRAPETALA, ULT1, ULTRAPETALA1 |
0.348 |
-0.003 |
| 293 |
262876_at |
AT1G64750
|
ATDSS1(I), deletion of SUV3 suppressor 1(I), DSS1(I), deletion of SUV3 suppressor 1(I) |
0.347 |
-0.082 |
| 294 |
255285_at |
AT4G04630
|
unknown |
0.347 |
-0.354 |
| 295 |
250809_at |
AT5G05140
|
[Transcription elongation factor (TFIIS) family protein] |
0.346 |
-0.300 |
| 296 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.345 |
-0.532 |
| 297 |
258468_at |
AT3G06070
|
unknown |
0.344 |
-0.815 |
| 298 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
0.343 |
-0.079 |
| 299 |
261938_at |
AT1G22510
|
[FUNCTIONS IN: zinc ion binding] |
0.343 |
0.044 |
| 300 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.342 |
-0.295 |
| 301 |
255016_at |
AT4G10120
|
ATSPS4F |
-1.060 |
-0.046 |
| 302 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.953 |
0.172 |
| 303 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.916 |
-0.181 |
| 304 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.874 |
-0.199 |
| 305 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.867 |
-0.432 |
| 306 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.829 |
0.067 |
| 307 |
252661_at |
AT3G44450
|
unknown |
-0.813 |
1.282 |
| 308 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.813 |
0.111 |
| 309 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.775 |
0.688 |
| 310 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.760 |
0.304 |
| 311 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.744 |
-0.049 |
| 312 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.744 |
0.869 |
| 313 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.737 |
0.809 |
| 314 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.734 |
0.183 |
| 315 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.725 |
0.123 |
| 316 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.720 |
0.035 |
| 317 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.719 |
0.496 |
| 318 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.716 |
0.745 |
| 319 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.714 |
0.213 |
| 320 |
252377_at |
AT3G47960
|
AtNPF2.10, GTR1, GLUCOSINOLATE TRANSPORTER-1, NPF2.10, NRT1/ PTR family 2.10 |
-0.711 |
0.203 |
| 321 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.702 |
-0.063 |
| 322 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.700 |
0.185 |
| 323 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.690 |
-0.094 |
| 324 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.683 |
-0.253 |
| 325 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
-0.678 |
-0.133 |
| 326 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.678 |
0.269 |
| 327 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.673 |
0.225 |
| 328 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.668 |
-0.361 |
| 329 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.665 |
-0.456 |
| 330 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.654 |
-0.163 |
| 331 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.652 |
-0.351 |
| 332 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.638 |
-0.190 |
| 333 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.638 |
-0.145 |
| 334 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.626 |
-0.519 |
| 335 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
-0.621 |
0.324 |
| 336 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.617 |
0.033 |
| 337 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.599 |
0.335 |
| 338 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.597 |
0.299 |
| 339 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.595 |
0.165 |
| 340 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.591 |
0.117 |
| 341 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.587 |
0.217 |
| 342 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.577 |
0.422 |
| 343 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.571 |
0.105 |
| 344 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.571 |
-0.102 |
| 345 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.570 |
0.182 |
| 346 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.569 |
0.523 |
| 347 |
256603_at |
AT3G28270
|
unknown |
-0.563 |
0.144 |
| 348 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.561 |
0.084 |
| 349 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.558 |
0.249 |
| 350 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
-0.558 |
0.467 |
| 351 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.554 |
0.675 |
| 352 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.550 |
0.074 |
| 353 |
247486_at |
AT5G62140
|
unknown |
-0.549 |
0.257 |
| 354 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.549 |
1.124 |
| 355 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.547 |
0.066 |
| 356 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.545 |
0.634 |
| 357 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.543 |
0.395 |
| 358 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.543 |
-0.797 |
| 359 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.541 |
0.737 |
| 360 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-0.540 |
-0.254 |
| 361 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.538 |
0.854 |
| 362 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.535 |
-0.009 |
| 363 |
264909_at |
AT2G17300
|
unknown |
-0.531 |
-0.236 |
| 364 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.530 |
0.194 |
| 365 |
252619_at |
AT3G45210
|
unknown |
-0.525 |
0.092 |
| 366 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-0.524 |
0.493 |
| 367 |
251142_at |
AT5G01015
|
unknown |
-0.518 |
0.306 |
| 368 |
248028_at |
AT5G55620
|
unknown |
-0.517 |
0.545 |
| 369 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
-0.515 |
0.535 |
| 370 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
-0.512 |
1.160 |
| 371 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.511 |
0.721 |
| 372 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.504 |
-0.229 |
| 373 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
-0.502 |
0.438 |
| 374 |
258621_at |
AT3G02830
|
PNT1, PENTA 1, ZFN1, zinc finger protein 1 |
-0.500 |
0.623 |
| 375 |
254145_at |
AT4G24700
|
unknown |
-0.497 |
0.322 |
| 376 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.495 |
-0.192 |
| 377 |
261758_at |
AT1G08250
|
ADT6, arogenate dehydratase 6, AtADT6, Arabidopsis thaliana arogenate dehydratase 6 |
-0.494 |
-0.147 |
| 378 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.490 |
0.310 |
| 379 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.489 |
-0.010 |
| 380 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.489 |
-0.175 |
| 381 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.487 |
0.330 |
| 382 |
247977_at |
AT5G56850
|
unknown |
-0.481 |
0.789 |
| 383 |
250985_at |
AT5G02830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.473 |
0.433 |
| 384 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.471 |
0.028 |
| 385 |
262533_at |
AT1G17090
|
unknown |
-0.468 |
0.079 |
| 386 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.468 |
0.623 |
| 387 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.463 |
-0.552 |
| 388 |
257314_at |
AT3G26590
|
[MATE efflux family protein] |
-0.463 |
0.105 |
| 389 |
263410_at |
AT2G04039
|
unknown |
-0.458 |
0.746 |
| 390 |
261221_at |
AT1G19960
|
unknown |
-0.457 |
-0.461 |
| 391 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.456 |
-0.001 |
| 392 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.453 |
-0.039 |
| 393 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.450 |
0.395 |
| 394 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.450 |
0.016 |
| 395 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
-0.449 |
0.015 |
| 396 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.448 |
0.596 |
| 397 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.446 |
0.187 |
| 398 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.445 |
1.044 |
| 399 |
259655_at |
AT1G55210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.439 |
0.023 |
| 400 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
-0.439 |
0.424 |
| 401 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
-0.438 |
-0.003 |
| 402 |
264444_at |
AT1G27360
|
SPL11, squamosa promoter-like 11 |
-0.437 |
0.243 |
| 403 |
263674_at |
AT2G04790
|
unknown |
-0.432 |
0.758 |
| 404 |
257533_at |
AT3G10840
|
[alpha/beta-Hydrolases superfamily protein] |
-0.430 |
0.412 |
| 405 |
247679_at |
AT5G59540
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.429 |
0.312 |
| 406 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.428 |
0.428 |
| 407 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.425 |
0.014 |
| 408 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.420 |
1.037 |
| 409 |
252888_at |
AT4G39210
|
APL3 |
-0.420 |
0.308 |
| 410 |
262811_at |
AT1G11700
|
unknown |
-0.420 |
-0.048 |
| 411 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.419 |
0.217 |
| 412 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.418 |
0.038 |
| 413 |
251759_at |
AT3G55630
|
ATDFD, DHFS-FPGS homolog D, DFD, DHFS-FPGS homolog D, FPGS3, folylpolyglutamate synthetase 3 |
-0.418 |
0.430 |
| 414 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-0.417 |
0.565 |
| 415 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.417 |
0.006 |
| 416 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.417 |
0.582 |
| 417 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-0.416 |
0.954 |
| 418 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.415 |
0.182 |
| 419 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.414 |
0.112 |
| 420 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.414 |
0.718 |
| 421 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.411 |
0.067 |
| 422 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.410 |
-0.486 |
| 423 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.408 |
0.027 |
| 424 |
267319_at |
AT2G34660
|
ABCC2, ATP-binding cassette C2, AtABCC2, Arabidopsis thaliana ATP-binding cassette C2, ATMRP2, multidrug resistance-associated protein 2, EST4, MRP2, multidrug resistance-associated protein 2 |
-0.407 |
0.252 |
| 425 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
-0.407 |
0.570 |
| 426 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-0.407 |
0.315 |
| 427 |
267294_at |
AT2G23670
|
YCF37, homolog of Synechocystis YCF37 |
-0.405 |
0.643 |
| 428 |
259275_at |
AT3G01060
|
unknown |
-0.401 |
0.582 |
| 429 |
265182_at |
AT1G23740
|
AOR, alkenal/one oxidoreductase |
-0.400 |
0.333 |
| 430 |
264201_at |
AT1G22630
|
unknown |
-0.400 |
0.659 |
| 431 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
-0.399 |
0.246 |
| 432 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.397 |
0.424 |
| 433 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.396 |
0.366 |
| 434 |
262162_at |
AT1G78020
|
unknown |
-0.394 |
0.016 |
| 435 |
262745_at |
AT1G28600
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.394 |
0.365 |
| 436 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.394 |
-0.490 |
| 437 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.393 |
0.723 |
| 438 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.393 |
0.299 |
| 439 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.392 |
0.509 |
| 440 |
265742_at |
AT2G01290
|
RPI2, ribose-5-phosphate isomerase 2 |
-0.391 |
0.448 |
| 441 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.390 |
0.864 |
| 442 |
250828_at |
AT5G05250
|
unknown |
-0.389 |
-0.126 |
| 443 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.388 |
0.480 |
| 444 |
254642_at |
AT4G18810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.388 |
0.588 |
| 445 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.387 |
0.427 |
| 446 |
253228_at |
AT4G34630
|
unknown |
-0.387 |
0.226 |
| 447 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.387 |
-0.131 |
| 448 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.387 |
0.041 |
| 449 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.386 |
-0.082 |
| 450 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
-0.386 |
0.297 |
| 451 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
-0.385 |
0.156 |
| 452 |
261684_at |
AT1G47400
|
unknown |
-0.385 |
-0.224 |
| 453 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.384 |
0.707 |
| 454 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.383 |
0.469 |
| 455 |
247214_at |
AT5G64850
|
unknown |
-0.383 |
0.427 |
| 456 |
247347_at |
AT5G63780
|
SHA1, shoot apical meristem arrest 1 |
-0.383 |
0.192 |
| 457 |
252010_at |
AT3G52740
|
unknown |
-0.383 |
1.067 |
| 458 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
-0.382 |
-0.610 |
| 459 |
255957_at |
AT1G22160
|
unknown |
-0.381 |
-0.403 |
| 460 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.380 |
0.176 |
| 461 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
-0.380 |
0.265 |
| 462 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.380 |
-0.065 |
| 463 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.379 |
0.225 |
| 464 |
250944_at |
AT5G03380
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.376 |
-0.026 |
| 465 |
245165_at |
AT2G33180
|
unknown |
-0.376 |
0.521 |
| 466 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.375 |
0.097 |
| 467 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.374 |
0.596 |
| 468 |
252353_at |
AT3G48200
|
unknown |
-0.373 |
0.416 |
| 469 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.373 |
0.721 |
| 470 |
260696_at |
AT1G32520
|
unknown |
-0.373 |
0.493 |
| 471 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.373 |
-0.020 |
| 472 |
264636_at |
AT1G65490
|
unknown |
-0.373 |
1.198 |
| 473 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.373 |
0.293 |
| 474 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.372 |
0.058 |
| 475 |
256186_at |
AT1G51680
|
4CL.1, 4-COUMARATE:COA LIGASE 1, 4CL1, 4-coumarate:CoA ligase 1, AT4CL1, ARABIDOPSIS THALIANA 4-COUMARATE:COA LIGASE 1 |
-0.372 |
-0.279 |
| 476 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.372 |
0.174 |
| 477 |
253922_at |
AT4G26850
|
VTC2, vitamin c defective 2 |
-0.371 |
0.184 |
| 478 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
-0.370 |
1.105 |
| 479 |
254133_at |
AT4G24810
|
[Protein kinase superfamily protein] |
-0.370 |
0.356 |
| 480 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.368 |
-0.016 |
| 481 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
-0.368 |
0.349 |
| 482 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.367 |
0.443 |
| 483 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
-0.366 |
0.374 |
| 484 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.364 |
0.125 |
| 485 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.364 |
-0.110 |
| 486 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.364 |
0.494 |
| 487 |
252073_at |
AT3G51750
|
unknown |
-0.364 |
0.507 |
| 488 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
-0.363 |
1.159 |
| 489 |
263599_at |
AT2G01830
|
AHK4, ARABIDOPSIS HISTIDINE KINASE 4, ATCRE1, CRE1, CYTOKININ RESPONSE 1, WOL, WOODEN LEG, WOL1, WOODEN LEG 1 |
-0.362 |
-0.441 |
| 490 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.361 |
0.065 |
| 491 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.361 |
0.394 |
| 492 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
-0.360 |
0.413 |
| 493 |
253387_at |
AT4G33010
|
AtGLDP1, glycine decarboxylase P-protein 1, GLDP1, glycine decarboxylase P-protein 1 |
-0.360 |
0.441 |
| 494 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
-0.359 |
0.115 |
| 495 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.358 |
-0.531 |
| 496 |
246445_at |
AT5G17630
|
[Nucleotide/sugar transporter family protein] |
-0.357 |
0.085 |
| 497 |
247235_at |
AT5G64580
|
EMB3144, EMBRYO DEFECTIVE 3144 |
-0.357 |
0.386 |
| 498 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.356 |
-0.269 |
| 499 |
258982_at |
AT3G08870
|
LecRK-VI.1, L-type lectin receptor kinase VI.1 |
-0.356 |
0.484 |
| 500 |
254431_at |
AT4G20840
|
[FAD-binding Berberine family protein] |
-0.355 |
-0.137 |
| 501 |
260387_at |
AT1G74100
|
ATSOT16, SULFOTRANSFERASE 16, ATST5A, ARABIDOPSIS SULFOTRANSFERASE 5A, CORI-7, CORONATINE INDUCED-7, SOT16, sulfotransferase 16 |
-0.354 |
-0.229 |
| 502 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-0.354 |
1.030 |
| 503 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
-0.352 |
0.465 |
| 504 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.351 |
0.223 |
| 505 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.350 |
0.574 |
| 506 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.350 |
0.185 |
| 507 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
-0.348 |
-0.201 |
| 508 |
266384_at |
AT2G14660
|
unknown |
-0.348 |
0.331 |
| 509 |
267299_at |
AT2G30150
|
[UDP-Glycosyltransferase superfamily protein] |
-0.348 |
0.408 |
| 510 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.346 |
0.630 |
| 511 |
250960_at |
AT5G02940
|
unknown |
-0.345 |
0.564 |
| 512 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.345 |
1.145 |
| 513 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.345 |
0.145 |
| 514 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
-0.345 |
-0.038 |
| 515 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.344 |
0.753 |
| 516 |
257880_at |
AT3G16910
|
AAE7, acyl-activating enzyme 7, ACN1, ACETATE NON-UTILIZING 1 |
-0.344 |
-0.113 |
| 517 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.343 |
0.357 |
| 518 |
262536_at |
AT1G17100
|
AtHBP1, HBP1, haem-binding protein 1 |
-0.343 |
0.568 |
| 519 |
253708_at |
AT4G29210
|
GGT3, gamma-glutamyl transpeptidase 3, GGT4, gamma-glutamyl transpeptidase 4 |
-0.343 |
-0.193 |
| 520 |
248582_at |
AT5G49910
|
cpHsc70-2, chloroplast heat shock protein 70-2, HSC70-7, HEAT SHOCK PROTEIN 70-7 |
-0.342 |
0.339 |
| 521 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.342 |
-0.473 |
| 522 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
-0.342 |
0.035 |
| 523 |
263714_at |
AT2G20610
|
ALF1, ABERRANT LATERAL ROOT FORMATION 1, HLS3, HOOKLESS 3, RTY, ROOTY, RTY1, ROOTY 1, SUR1, SUPERROOT 1 |
-0.341 |
-0.167 |
| 524 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
-0.341 |
0.178 |
| 525 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.340 |
0.038 |
| 526 |
245296_at |
AT4G16370
|
oligopeptide transporter |
-0.340 |
0.221 |
| 527 |
247400_at |
AT5G62840
|
[Phosphoglycerate mutase family protein] |
-0.340 |
0.668 |
| 528 |
251594_at |
AT3G57630
|
[exostosin family protein] |
-0.339 |
-0.100 |
| 529 |
256418_at |
AT3G06160
|
[AP2/B3-like transcriptional factor family protein] |
-0.339 |
-0.007 |
| 530 |
256892_at |
AT3G19000
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.338 |
0.095 |
| 531 |
250429_at |
AT5G10470
|
KAC1, kinesin like protein for actin based chloroplast movement 1, KCA1, KINESIN CDKA;1 ASSOCIATED 1 |
-0.338 |
0.581 |
| 532 |
261921_at |
AT1G65900
|
unknown |
-0.337 |
0.191 |
| 533 |
245936_at |
AT5G19850
|
[alpha/beta-Hydrolases superfamily protein] |
-0.337 |
0.630 |
| 534 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.336 |
0.765 |
| 535 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.336 |
0.076 |
| 536 |
264873_at |
AT1G24100
|
UGT74B1, UDP-glucosyl transferase 74B1 |
-0.335 |
-0.206 |
| 537 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.334 |
0.181 |
| 538 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.334 |
-0.450 |
| 539 |
264002_at |
AT2G22360
|
DJA6, DNA J protein A6 |
-0.334 |
0.326 |
| 540 |
257706_at |
AT3G12685
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
-0.334 |
0.279 |
| 541 |
258179_at |
AT3G21690
|
[MATE efflux family protein] |
-0.333 |
0.315 |
| 542 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.333 |
0.105 |
| 543 |
258023_at |
AT3G19450
|
ATCAD4, CAD, CAD-C, CAD4, CINNAMYL ALCOHOL DEHYDROGENASE 4 |
-0.333 |
0.168 |
| 544 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.332 |
0.021 |
| 545 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.332 |
-0.560 |
| 546 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.332 |
-0.633 |
| 547 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.331 |
0.005 |
| 548 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
-0.331 |
0.149 |
| 549 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.331 |
0.215 |
| 550 |
260385_at |
AT1G74090
|
ATSOT18, DESULFO-GLUCOSINOLATE SULFOTRANSFERASE 18, ATST5B, ARABIDOPSIS SULFOTRANSFERASE 5B, SOT18, desulfo-glucosinolate sulfotransferase 18 |
-0.330 |
-0.085 |
| 551 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
-0.330 |
0.127 |
| 552 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.330 |
-0.740 |
| 553 |
264963_at |
AT1G60600
|
ABC4, ABERRANT CHLOROPLAST DEVELOPMENT 4 |
-0.329 |
0.273 |
| 554 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.328 |
-0.222 |
| 555 |
263477_at |
AT2G31790
|
[UDP-Glycosyltransferase superfamily protein] |
-0.327 |
0.300 |
| 556 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.327 |
0.341 |
| 557 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.326 |
0.464 |
| 558 |
263533_at |
AT2G24820
|
AtTic55, translocon at the inner envelope membrane of chloroplasts 55, Tic55, translocon at the inner envelope membrane of chloroplasts 55, TIC55-II, translocon at the inner envelope membrane of chloroplasts 55-II |
-0.326 |
0.313 |
| 559 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.326 |
-0.535 |
| 560 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
-0.325 |
0.320 |
| 561 |
254693_at |
AT4G17880
|
MYC4 |
-0.325 |
-0.303 |
| 562 |
245368_at |
AT4G15510
|
PPD1, PsbP-Domain Protein1 |
-0.324 |
0.117 |
| 563 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
-0.324 |
0.119 |
| 564 |
246976_s_at |
AT5G24810
|
[ABC1 family protein] |
-0.324 |
0.267 |
| 565 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
-0.323 |
0.833 |
| 566 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.323 |
0.563 |
| 567 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.322 |
-0.005 |
| 568 |
248634_at |
AT5G49030
|
OVA2, ovule abortion 2 |
-0.322 |
0.309 |
| 569 |
248853_at |
AT5G46570
|
BSK2, brassinosteroid-signaling kinase 2 |
-0.322 |
0.085 |
| 570 |
256617_at |
AT3G22240
|
unknown |
-0.322 |
-0.074 |
| 571 |
258609_at |
AT3G02910
|
[AIG2-like (avirulence induced gene) family protein] |
-0.321 |
0.257 |
| 572 |
255676_at |
AT4G00490
|
BAM2, beta-amylase 2, BMY9, BETA-AMYLASE 9 |
-0.321 |
0.217 |
| 573 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
-0.321 |
0.111 |
| 574 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.320 |
-0.063 |
| 575 |
256674_at |
AT3G52360
|
unknown |
-0.320 |
-0.291 |
| 576 |
267380_at |
AT2G26170
|
CYP711A1, cytochrome P450, family 711, subfamily A, polypeptide 1, MAX1, MORE AXILLARY BRANCHES 1 |
-0.320 |
0.033 |
| 577 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.319 |
0.024 |
| 578 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.318 |
0.163 |
| 579 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
-0.318 |
0.001 |
| 580 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
-0.318 |
0.033 |
| 581 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.318 |
-0.431 |
| 582 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.317 |
0.151 |
| 583 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.317 |
0.190 |
| 584 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.316 |
0.016 |
| 585 |
248201_at |
AT5G54180
|
PTAC15, plastid transcriptionally active 15 |
-0.316 |
0.461 |
| 586 |
266338_at |
AT2G32400
|
ATGLR3.7, GLR3.7, GLUTAMATE RECEPTOR 3.7, GLR5, glutamate receptor 5 |
-0.314 |
0.392 |
| 587 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
-0.314 |
0.160 |
| 588 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.313 |
0.088 |
| 589 |
267112_at |
AT2G14750
|
AKN1, APS KINASE 1, APK, APS kinase, APK1, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 1, ATAKN1 |
-0.313 |
-0.023 |
| 590 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.313 |
0.170 |
| 591 |
261801_at |
AT1G30520
|
AAE14, acyl-activating enzyme 14 |
-0.312 |
0.157 |
| 592 |
253302_at |
AT4G33660
|
unknown |
-0.312 |
0.338 |
| 593 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
-0.312 |
0.063 |
| 594 |
260237_at |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
-0.311 |
0.401 |
| 595 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
-0.310 |
0.298 |
| 596 |
266882_at |
AT2G44670
|
unknown |
-0.310 |
0.001 |
| 597 |
247693_at |
AT5G59730
|
ATEXO70H7, exocyst subunit exo70 family protein H7, EXO70H7, exocyst subunit exo70 family protein H7 |
-0.309 |
0.105 |
| 598 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.308 |
-0.487 |
| 599 |
248865_at |
AT5G46790
|
PYL1, PYR1-like 1, RCAR12, regulatory components of ABA receptor 12 |
-0.308 |
-0.202 |
| 600 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.308 |
-0.940 |