|
probeID |
AGICode |
Annotation |
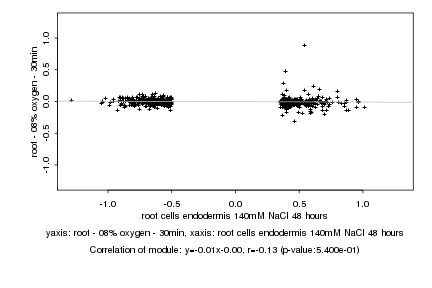
Log2 signal ratio
root cells endodermis 140mM NaCl 48 hours |
Log2 signal ratio
root - 08% oxygen - 30min |
| 1 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
1.011 |
-0.086 |
| 2 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
0.964 |
0.015 |
| 3 |
253070_at |
AT4G37850
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.964 |
-0.015 |
| 4 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.946 |
-0.080 |
| 5 |
253503_at |
AT4G31950
|
CYP82C3, cytochrome P450, family 82, subfamily C, polypeptide 3 |
0.942 |
0.039 |
| 6 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.883 |
-0.127 |
| 7 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.871 |
-0.131 |
| 8 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
0.866 |
-0.003 |
| 9 |
259453_at |
AT1G44090
|
ATGA20OX5, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 5, GA20OX5, gibberellin 20-oxidase 5 |
0.865 |
0.026 |
| 10 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.854 |
-0.069 |
| 11 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.851 |
-0.029 |
| 12 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.831 |
-0.025 |
| 13 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.807 |
-0.010 |
| 14 |
257026_at |
AT3G19200
|
unknown |
0.800 |
0.077 |
| 15 |
263481_at |
AT2G04025
|
GLV7, GOLVEN 7, RGF3, root meristem growth factor 3 |
0.793 |
0.164 |
| 16 |
252364_at |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.754 |
-0.007 |
| 17 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
0.736 |
0.058 |
| 18 |
256125_at |
AT1G18250
|
ATLP-1 |
0.731 |
-0.035 |
| 19 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.728 |
-0.065 |
| 20 |
260528_at |
AT2G47260
|
ATWRKY23, WRKY DNA-BINDING PROTEIN 23, WRKY23, WRKY DNA-binding protein 23 |
0.719 |
-0.004 |
| 21 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.713 |
-0.138 |
| 22 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
0.710 |
0.032 |
| 23 |
246103_at |
AT5G28640
|
AN3, ANGUSTIFOLIA 3, ATGIF1, ARABIDOPSIS GRF1-INTERACTING FACTOR 1, GIF, GRF1-INTERACTING FACTOR, GIF1, GRF1-INTERACTING FACTOR 1 |
0.704 |
0.013 |
| 24 |
259129_at |
AT3G02150
|
PTF1, plastid transcription factor 1, TCP13, TEOSINTE BRANCHED1, CYCLOIDEA AND PCF TRANSCRIPTION FACTOR 13, TFPD |
0.699 |
-0.004 |
| 25 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.697 |
-0.037 |
| 26 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.697 |
-0.190 |
| 27 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
0.685 |
-0.023 |
| 28 |
260883_at |
AT1G29270
|
unknown |
0.682 |
-0.064 |
| 29 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
0.677 |
0.064 |
| 30 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.676 |
-0.134 |
| 31 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.675 |
-0.013 |
| 32 |
264718_at |
AT1G70130
|
LecRK-V.2, L-type lectin receptor kinase V.2 |
0.674 |
-0.027 |
| 33 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
0.670 |
0.015 |
| 34 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
0.663 |
0.003 |
| 35 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
0.663 |
0.005 |
| 36 |
252739_at |
AT3G43250
|
unknown |
0.661 |
-0.011 |
| 37 |
260668_at |
AT1G19530
|
unknown |
0.658 |
0.192 |
| 38 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
0.654 |
0.003 |
| 39 |
256710_at |
AT3G30350
|
GLV3, GOLVEN 3, RGF4, root meristem growth factor 4 |
0.648 |
0.080 |
| 40 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.642 |
-0.041 |
| 41 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.641 |
0.048 |
| 42 |
250426_at |
AT5G10510
|
AIL6, AINTEGUMENTA-like 6, PLT3, PLETHORA 3 |
0.632 |
0.020 |
| 43 |
266223_at |
AT2G28790
|
[Pathogenesis-related thaumatin superfamily protein] |
0.626 |
-0.029 |
| 44 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.624 |
-0.083 |
| 45 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
0.621 |
0.036 |
| 46 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.614 |
-0.016 |
| 47 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
0.611 |
0.022 |
| 48 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
0.609 |
-0.012 |
| 49 |
246463_at |
AT5G16970
|
AER, alkenal reductase, AT-AER, alkenal reductase |
0.607 |
-0.029 |
| 50 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.605 |
0.244 |
| 51 |
245674_at |
AT1G56680
|
[Chitinase family protein] |
0.604 |
-0.077 |
| 52 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
0.601 |
0.049 |
| 53 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.594 |
-0.003 |
| 54 |
255782_at |
AT1G19850
|
ARF5, AUXIN RESPONSE FACTOR 5, IAA24, indole-3-acetic acid inducible 24, MP, MONOPTEROS |
0.591 |
0.003 |
| 55 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
0.591 |
-0.163 |
| 56 |
264774_at |
AT1G22890
|
unknown |
0.590 |
-0.049 |
| 57 |
256712_at |
AT2G34020
|
[Calcium-binding EF-hand family protein] |
0.589 |
-0.049 |
| 58 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.588 |
-0.152 |
| 59 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.586 |
-0.120 |
| 60 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
0.584 |
0.002 |
| 61 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
0.583 |
-0.175 |
| 62 |
258397_at |
AT3G15357
|
unknown |
0.577 |
-0.007 |
| 63 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.576 |
0.046 |
| 64 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
0.576 |
0.002 |
| 65 |
262314_at |
AT1G70810
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.575 |
-0.021 |
| 66 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.574 |
0.021 |
| 67 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.572 |
-0.033 |
| 68 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
0.572 |
-0.005 |
| 69 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
0.570 |
-0.073 |
| 70 |
263436_at |
AT2G28690
|
unknown |
0.569 |
-0.011 |
| 71 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.569 |
-0.039 |
| 72 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
0.568 |
-0.030 |
| 73 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.567 |
0.015 |
| 74 |
247061_at |
AT5G66780
|
unknown |
0.566 |
0.121 |
| 75 |
266455_at |
AT2G22760
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.562 |
-0.022 |
| 76 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
0.560 |
-0.062 |
| 77 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.557 |
-0.066 |
| 78 |
266860_at |
AT2G26870
|
NPC2, non-specific phospholipase C2 |
0.555 |
0.021 |
| 79 |
245944_at |
AT5G19520
|
ATMSL9, MSL9, mechanosensitive channel of small conductance-like 9 |
0.554 |
-0.017 |
| 80 |
260368_at |
AT1G69700
|
ATHVA22C, HVA22 homologue C, HVA22C, HVA22 homologue C |
0.552 |
-0.006 |
| 81 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.552 |
-0.025 |
| 82 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
0.551 |
-0.015 |
| 83 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
0.549 |
0.000 |
| 84 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.549 |
0.006 |
| 85 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
0.548 |
-0.026 |
| 86 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.547 |
-0.061 |
| 87 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.545 |
-0.025 |
| 88 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.541 |
0.897 |
| 89 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.538 |
0.182 |
| 90 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.534 |
-0.045 |
| 91 |
261921_at |
AT1G65900
|
unknown |
0.533 |
-0.023 |
| 92 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.526 |
0.004 |
| 93 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
0.525 |
-0.174 |
| 94 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
0.523 |
-0.000 |
| 95 |
248215_at |
AT5G53680
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.519 |
-0.010 |
| 96 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.519 |
-0.026 |
| 97 |
251807_at |
AT3G55400
|
OVA1, OVULE ABORTION 1 |
0.518 |
0.010 |
| 98 |
253299_at |
AT4G33800
|
unknown |
0.516 |
0.031 |
| 99 |
251456_at |
AT3G60120
|
BGLU27, beta glucosidase 27 |
0.515 |
-0.005 |
| 100 |
246212_at |
AT4G36930
|
SPT, SPATULA |
0.514 |
0.042 |
| 101 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.511 |
0.047 |
| 102 |
252077_at |
AT3G51720
|
unknown |
0.511 |
-0.014 |
| 103 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.510 |
-0.038 |
| 104 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.509 |
-0.033 |
| 105 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.503 |
-0.003 |
| 106 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.503 |
0.066 |
| 107 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
0.502 |
0.026 |
| 108 |
260974_at |
AT1G53440
|
[Leucine-rich repeat transmembrane protein kinase] |
0.501 |
-0.086 |
| 109 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.501 |
-0.021 |
| 110 |
257191_at |
AT3G13175
|
unknown |
0.499 |
-0.093 |
| 111 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.498 |
0.023 |
| 112 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
0.497 |
0.007 |
| 113 |
251355_at |
AT3G61100
|
[Putative endonuclease or glycosyl hydrolase] |
0.496 |
-0.010 |
| 114 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.492 |
-0.163 |
| 115 |
246418_at |
AT5G16960
|
[Zinc-binding dehydrogenase family protein] |
0.491 |
-0.002 |
| 116 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
0.491 |
-0.014 |
| 117 |
260688_at |
AT1G17665
|
unknown |
0.491 |
-0.062 |
| 118 |
245352_at |
AT4G15490
|
UGT84A3 |
0.490 |
-0.044 |
| 119 |
259363_at |
AT1G13270
|
MAP1B, METHIONINE AMINOPEPTIDASE 1B, MAP1C, methionine aminopeptidase 1B |
0.487 |
-0.070 |
| 120 |
253632_at |
AT4G30430
|
TET9, tetraspanin9 |
0.486 |
-0.025 |
| 121 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.486 |
-0.004 |
| 122 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
0.485 |
0.017 |
| 123 |
247268_at |
AT5G64080
|
AtXYP1, XYP1, xylogen protein 1 |
0.484 |
-0.007 |
| 124 |
257074_at |
AT3G19660
|
unknown |
0.482 |
0.023 |
| 125 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.482 |
-0.013 |
| 126 |
263881_at |
AT2G21820
|
unknown |
0.480 |
0.034 |
| 127 |
259225_at |
AT3G07590
|
[Small nuclear ribonucleoprotein family protein] |
0.480 |
-0.018 |
| 128 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.477 |
-0.006 |
| 129 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.476 |
-0.052 |
| 130 |
253740_at |
AT4G28706
|
[pfkB-like carbohydrate kinase family protein] |
0.474 |
-0.028 |
| 131 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.474 |
0.005 |
| 132 |
260535_at |
AT2G43390
|
unknown |
0.474 |
0.018 |
| 133 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
0.474 |
0.017 |
| 134 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
0.474 |
0.000 |
| 135 |
249060_at |
AT5G44560
|
VPS2.2 |
0.472 |
-0.003 |
| 136 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.472 |
0.038 |
| 137 |
261941_at |
AT1G22490
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.469 |
-0.012 |
| 138 |
248626_at |
AT5G48940
|
[Leucine-rich repeat transmembrane protein kinase family protein] |
0.468 |
0.024 |
| 139 |
255094_at |
AT4G08590
|
ORL1, ORTH-LIKE 1, ORL1, ORTHRUS-LIKE 1, ORTHL, ORTHRUS-like, VIM6, VARIANT IN METHYLATION 6 |
0.467 |
0.011 |
| 140 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.465 |
0.043 |
| 141 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.464 |
0.016 |
| 142 |
250622_at |
AT5G07310
|
[Integrase-type DNA-binding superfamily protein] |
0.463 |
0.022 |
| 143 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.461 |
0.047 |
| 144 |
258108_at |
AT3G23570
|
[alpha/beta-Hydrolases superfamily protein] |
0.460 |
0.004 |
| 145 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.460 |
-0.062 |
| 146 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
0.460 |
0.021 |
| 147 |
259823_at |
AT1G66250
|
[O-Glycosyl hydrolases family 17 protein] |
0.458 |
0.005 |
| 148 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.457 |
-0.035 |
| 149 |
248316_at |
AT5G52670
|
[Copper transport protein family] |
0.457 |
-0.305 |
| 150 |
248696_at |
AT5G48360
|
[Actin-binding FH2 (formin homology 2) family protein] |
0.457 |
0.022 |
| 151 |
264862_at |
AT1G24330
|
[ARM repeat superfamily protein] |
0.456 |
0.024 |
| 152 |
265395_at |
AT2G20850
|
SRF1, STRUBBELIG-receptor family 1 |
0.456 |
0.002 |
| 153 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
0.456 |
-0.008 |
| 154 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.455 |
-0.037 |
| 155 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.454 |
0.000 |
| 156 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
0.452 |
0.026 |
| 157 |
251050_at |
AT5G02440
|
unknown |
0.451 |
0.013 |
| 158 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.451 |
-0.029 |
| 159 |
246370_at |
AT1G51920
|
unknown |
0.450 |
-0.061 |
| 160 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.449 |
-0.009 |
| 161 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
0.446 |
-0.038 |
| 162 |
255945_at |
AT5G28610
|
unknown |
0.444 |
0.024 |
| 163 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.441 |
-0.003 |
| 164 |
256763_at |
AT3G16860
|
COBL8, COBRA-like protein 8 precursor |
0.438 |
0.002 |
| 165 |
267207_at |
AT2G30840
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.436 |
0.015 |
| 166 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.436 |
0.029 |
| 167 |
257353_at |
AT2G23050
|
MEL4, MAB4/ENP/NPY1-LIKE 4, NPY4, NAKED PINS IN YUC MUTANTS 4 |
0.435 |
-0.009 |
| 168 |
249747_at |
AT5G24600
|
unknown |
0.435 |
-0.025 |
| 169 |
258599_at |
AT3G04520
|
THA2, threonine aldolase 2 |
0.434 |
0.009 |
| 170 |
246274_at |
AT4G36620
|
GATA19, GATA transcription factor 19, HANL2, hanaba taranu like 2 |
0.434 |
-0.008 |
| 171 |
263120_at |
AT1G78490
|
CYP708A3, cytochrome P450, family 708, subfamily A, polypeptide 3 |
0.434 |
-0.039 |
| 172 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.431 |
-0.082 |
| 173 |
257879_at |
AT3G17160
|
unknown |
0.431 |
-0.006 |
| 174 |
252212_at |
AT3G50310
|
MAPKKK20, mitogen-activated protein kinase kinase kinase 20, MKKK20, MAPKK kinase 20 |
0.431 |
-0.071 |
| 175 |
247622_at |
AT5G60350
|
unknown |
0.431 |
-0.022 |
| 176 |
251882_at |
AT3G54140
|
AtNPF8.1, ATPTR1, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 1, NPF8.1, NRT1/ PTR family 8.1, PTR1, peptide transporter 1 |
0.430 |
-0.026 |
| 177 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
0.430 |
0.014 |
| 178 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.430 |
-0.054 |
| 179 |
254459_at |
AT4G21200
|
ATGA2OX8, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 8, GA2OX8, gibberellin 2-oxidase 8 |
0.428 |
-0.021 |
| 180 |
264799_at |
AT1G08550
|
AVDE1, ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1, NPQ1, non-photochemical quenching 1 |
0.428 |
-0.017 |
| 181 |
250695_at |
AT5G06740
|
LecRK-S.5, L-type lectin receptor kinase S.5 |
0.428 |
-0.039 |
| 182 |
264529_at |
AT1G30820
|
[CTP synthase family protein] |
0.428 |
-0.007 |
| 183 |
252282_at |
AT3G49360
|
PGL2, 6-phosphogluconolactonase 2 |
0.427 |
0.035 |
| 184 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
0.426 |
0.007 |
| 185 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.426 |
0.046 |
| 186 |
252543_at |
AT3G45780
|
JK224, NPH1, NONPHOTOTROPIC HYPOCOTYL 1, PHOT1, phototropin 1, RPT1, ROOT PHOTOTROPISM 1 |
0.424 |
-0.002 |
| 187 |
245828_at |
AT1G57820
|
ORTH2, ORTHRUS 2, VIM1, VARIANT IN METHYLATION 1 |
0.423 |
0.029 |
| 188 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.422 |
-0.036 |
| 189 |
250538_at |
AT5G08620
|
ATRH25, RNA HELICASE 25, STRS2, STRESS RESPONSE SUPPRESSOR 2 |
0.422 |
0.001 |
| 190 |
245488_at |
AT4G16270
|
[Peroxidase superfamily protein] |
0.422 |
0.065 |
| 191 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
0.422 |
-0.001 |
| 192 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.421 |
-0.019 |
| 193 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
0.421 |
0.026 |
| 194 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
0.421 |
-0.005 |
| 195 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
0.419 |
-0.010 |
| 196 |
249427_at |
AT5G39850
|
[Ribosomal protein S4] |
0.419 |
-0.006 |
| 197 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
0.418 |
-0.024 |
| 198 |
263360_at |
AT2G03830
|
GLV6, GOLVEN 6, RGF8, root meristem growth factor 8 |
0.418 |
0.006 |
| 199 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.416 |
0.058 |
| 200 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
0.414 |
-0.007 |
| 201 |
260051_at |
AT1G78210
|
[alpha/beta-Hydrolases superfamily protein] |
0.414 |
-0.003 |
| 202 |
262605_at |
AT1G15170
|
[MATE efflux family protein] |
0.414 |
0.020 |
| 203 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.413 |
0.026 |
| 204 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.413 |
0.011 |
| 205 |
246335_at |
AT3G44880
|
ACD1, ACCELERATED CELL DEATH 1, LLS1, LETHAL LEAF-SPOT 1 HOMOLOG, PAO, PHEOPHORBIDE A OXYGENASE |
0.412 |
-0.104 |
| 206 |
251667_at |
AT3G57150
|
AtCBF5, AtNAP57, CBF5, NAP57, homologue of NAP57 |
0.412 |
0.009 |
| 207 |
246426_at |
AT5G17430
|
BBM, BABY BOOM |
0.412 |
0.033 |
| 208 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.411 |
0.052 |
| 209 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
0.411 |
-0.021 |
| 210 |
250546_at |
AT5G08180
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.410 |
0.016 |
| 211 |
258527_at |
AT3G06850
|
BCE2, DIN3, DARK INDUCIBLE 3, LTA1 |
0.409 |
-0.070 |
| 212 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.409 |
0.011 |
| 213 |
252073_at |
AT3G51750
|
unknown |
0.409 |
0.058 |
| 214 |
262809_at |
AT1G11720
|
ATSS3, starch synthase 3, SS3, starch synthase 3 |
0.407 |
-0.004 |
| 215 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
0.405 |
-0.013 |
| 216 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.405 |
0.000 |
| 217 |
259008_at |
AT3G09390
|
ATMT-1, ARABIDOPSIS THALIANA METALLOTHIONEIN-1, ATMT-K, ARABIDOPSIS THALIANA METALLOTHIONEIN-K, MT2A, metallothionein 2A |
0.402 |
-0.060 |
| 218 |
257663_at |
AT3G20260
|
[FUNCTIONS IN: structural constituent of ribosome] |
0.402 |
0.004 |
| 219 |
252539_at |
AT3G45730
|
unknown |
0.401 |
-0.070 |
| 220 |
248613_at |
AT5G49555
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.401 |
0.006 |
| 221 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.401 |
-0.018 |
| 222 |
254522_at |
AT4G19980
|
unknown |
0.400 |
0.176 |
| 223 |
247671_at |
AT5G60210
|
RIP5, ROP interactive partner 5 |
0.400 |
0.036 |
| 224 |
265464_at |
AT2G37080
|
RIP2, ROP interactive partner 2 |
0.399 |
0.028 |
| 225 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
0.399 |
-0.021 |
| 226 |
245033_at |
AT2G26380
|
[Leucine-rich repeat (LRR) family protein] |
0.398 |
0.029 |
| 227 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.398 |
0.018 |
| 228 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.397 |
-0.162 |
| 229 |
248358_at |
AT5G52400
|
CYP715A1, cytochrome P450, family 715, subfamily A, polypeptide 1 |
0.397 |
-0.037 |
| 230 |
248660_at |
AT5G48650
|
[Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain] |
0.396 |
0.020 |
| 231 |
266658_at |
AT2G25735
|
unknown |
0.395 |
-0.117 |
| 232 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.395 |
0.012 |
| 233 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.394 |
-0.108 |
| 234 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
0.394 |
-0.003 |
| 235 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.393 |
-0.004 |
| 236 |
250532_at |
AT5G08540
|
unknown |
0.393 |
-0.017 |
| 237 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
0.393 |
-0.027 |
| 238 |
250152_at |
AT5G15120
|
unknown |
0.392 |
0.487 |
| 239 |
256780_at |
AT3G13640
|
ABCE1, ATP-binding cassette E1, ATRLI1, RNAse l inhibitor protein 1, RLI1, RNAse l inhibitor protein 1 |
0.392 |
0.034 |
| 240 |
260565_at |
AT2G43800
|
[Actin-binding FH2 (formin homology 2) family protein] |
0.387 |
0.035 |
| 241 |
267309_at |
AT2G19385
|
[zinc ion binding] |
0.387 |
0.018 |
| 242 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
0.386 |
-0.022 |
| 243 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.385 |
-0.059 |
| 244 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.385 |
-0.101 |
| 245 |
247813_at |
AT5G58330
|
[lactate/malate dehydrogenase family protein] |
0.384 |
-0.077 |
| 246 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.383 |
-0.015 |
| 247 |
262875_at |
AT1G64970
|
G-TMT, gamma-tocopherol methyltransferase, TMT1, VTE4, VITAMIN E DEFICIENT 4 |
0.382 |
0.066 |
| 248 |
249592_at |
AT5G37890
|
[Protein with RING/U-box and TRAF-like domains] |
0.382 |
0.049 |
| 249 |
256650_at |
AT3G13620
|
PUT4, POLYAMINE UPTAKE TRANSPORTER 4 |
0.382 |
0.040 |
| 250 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.381 |
0.064 |
| 251 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.381 |
-0.016 |
| 252 |
254831_at |
AT4G12600
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.380 |
-0.009 |
| 253 |
257718_at |
AT3G18400
|
anac058, NAC domain containing protein 58, NAC058, NAC domain containing protein 58 |
0.379 |
0.109 |
| 254 |
250738_at |
AT5G05730
|
AMT1, A-METHYL TRYPTOPHAN RESISTANT 1, ASA1, anthranilate synthase alpha subunit 1, JDL1, JASMONATE-INDUCED DEFECTIVE LATERAL ROOT 1, TRP5, TRYPTOPHAN BIOSYNTHESIS 5, WEI2, WEAK ETHYLENE INSENSITIVE 2 |
0.379 |
-0.045 |
| 255 |
266076_at |
AT2G40700
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.378 |
0.033 |
| 256 |
266018_at |
AT2G18710
|
SCY1, SECY homolog 1 |
0.378 |
0.001 |
| 257 |
260411_at |
AT1G69890
|
unknown |
0.376 |
-0.034 |
| 258 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
0.376 |
-0.012 |
| 259 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
0.376 |
0.045 |
| 260 |
256715_at |
AT2G34090
|
MEE18, maternal effect embryo arrest 18 |
0.376 |
-0.049 |
| 261 |
245571_at |
AT4G14695
|
[Uncharacterised protein family (UPF0041)] |
0.376 |
-0.017 |
| 262 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.374 |
0.288 |
| 263 |
254924_at |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
0.373 |
-0.010 |
| 264 |
251152_at |
AT3G63130
|
ATRANGAP1, RAN GTPASE-ACTIVATING PROTEIN 1, RANGAP1, RAN GTPase activating protein 1 |
0.373 |
-0.017 |
| 265 |
254504_at |
AT4G20030
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.371 |
-0.025 |
| 266 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
0.371 |
-0.024 |
| 267 |
259760_at |
AT1G77580
|
unknown |
0.370 |
0.056 |
| 268 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.369 |
-0.091 |
| 269 |
258257_at |
AT3G26770
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.368 |
0.006 |
| 270 |
247465_at |
AT5G62190
|
PRH75 |
0.368 |
0.001 |
| 271 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.367 |
-0.212 |
| 272 |
255888_at |
AT1G20300
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.367 |
-0.010 |
| 273 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
0.366 |
-0.047 |
| 274 |
260157_at |
AT1G52930
|
[Ribosomal RNA processing Brix domain protein] |
0.366 |
-0.005 |
| 275 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.365 |
-0.015 |
| 276 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
0.365 |
0.120 |
| 277 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.363 |
-0.001 |
| 278 |
262230_at |
AT1G68560
|
ATXYL1, ALPHA-XYLOSIDASE 1, AXY3, altered xyloglucan 3, TRG1, thermoinhibition resistant germination 1, XYL1, alpha-xylosidase 1 |
0.363 |
-0.018 |
| 279 |
262802_at |
AT1G20930
|
CDKB2;2, cyclin-dependent kinase B2;2 |
0.363 |
-0.013 |
| 280 |
263973_at |
AT2G42740
|
RPL16A, ribosomal protein large subunit 16A |
0.363 |
-0.020 |
| 281 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
0.363 |
-0.010 |
| 282 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.363 |
-0.013 |
| 283 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
0.363 |
-0.001 |
| 284 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.363 |
0.031 |
| 285 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
0.362 |
-0.007 |
| 286 |
245426_at |
AT4G17540
|
unknown |
0.362 |
0.035 |
| 287 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
0.362 |
0.034 |
| 288 |
246415_at |
AT5G17160
|
unknown |
0.362 |
0.003 |
| 289 |
253242_at |
AT4G34540
|
[NmrA-like negative transcriptional regulator family protein] |
0.361 |
0.022 |
| 290 |
264232_at |
AT1G67470
|
[Protein kinase superfamily protein] |
0.360 |
-0.040 |
| 291 |
261545_at |
AT1G63530
|
unknown |
0.358 |
0.019 |
| 292 |
265961_at |
AT2G37400
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.358 |
-0.044 |
| 293 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
0.356 |
-0.006 |
| 294 |
251797_at |
AT3G55560
|
AGF2, AT-hook protein of GA feedback 2, AHL15, AT-hook motif nuclear-localized protein 15 |
0.355 |
-0.012 |
| 295 |
248597_at |
AT5G49160
|
DDM2, DECREASED DNA METHYLATION 2, DMT01, DNA METHYLTRANSFERASE 01, DMT1, DNA METHYLTRANSFERASE 1, MET1, methyltransferase 1, MET2, METHYLTRANSFERASE 2, METI, METHYLTRANSFERASE I |
0.354 |
-0.013 |
| 296 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
0.354 |
-0.020 |
| 297 |
245800_at |
AT1G46264
|
AT-HSFB4, HSFB4, heat shock transcription factor B4, SCZ, SCHIZORIZA |
0.353 |
-0.002 |
| 298 |
261558_at |
AT1G01770
|
unknown |
0.353 |
0.014 |
| 299 |
255599_at |
AT4G01010
|
ATCNGC13, CYCLIC NUCLEOTIDE-GATED CHANNEL 13, CNGC13, cyclic nucleotide-gated channel 13 |
0.353 |
0.001 |
| 300 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.353 |
-0.079 |
| 301 |
256811_at |
AT3G21340
|
[Leucine-rich repeat protein kinase family protein] |
-1.292 |
0.023 |
| 302 |
267240_at |
AT2G02680
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.055 |
-0.026 |
| 303 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-1.049 |
0.013 |
| 304 |
246911_at |
AT5G25810
|
tny, TINY |
-1.045 |
-0.001 |
| 305 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-1.026 |
0.063 |
| 306 |
267228_at |
AT2G43890
|
[Pectin lyase-like superfamily protein] |
-0.994 |
-0.051 |
| 307 |
266913_at |
AT2G45890
|
ATROPGEF4, RHO GUANYL-NUCLEOTIDE EXCHANGE FACTOR 4, GEF4, guanine exchange factor 4, RHS11, ROOT HAIR SPECIFIC 11, ROPGEF4, ROP (rho of plants) guanine nucleotide exchange factor 4 |
-0.986 |
-0.015 |
| 308 |
261550_at |
AT1G63450
|
RHS8, root hair specific 8, XUT1, XYLOGLUCAN-SPECIFIC GALACTURONOSYLTRANSFERASE 1 |
-0.964 |
0.007 |
| 309 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
-0.961 |
0.036 |
| 310 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.930 |
-0.132 |
| 311 |
251832_at |
AT3G55150
|
ATEXO70H1, exocyst subunit exo70 family protein H1, EXO70H1, exocyst subunit exo70 family protein H1 |
-0.920 |
0.001 |
| 312 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.913 |
0.075 |
| 313 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
-0.908 |
0.076 |
| 314 |
250173_at |
AT5G14340
|
AtMYB40, myb domain protein 40, MYB40, myb domain protein 40 |
-0.907 |
0.023 |
| 315 |
263616_at |
AT2G04680
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.904 |
0.019 |
| 316 |
261647_at |
AT1G27740
|
RSL4, root hair defective 6-like 4 |
-0.904 |
0.054 |
| 317 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.902 |
-0.056 |
| 318 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
-0.900 |
-0.019 |
| 319 |
257405_at |
AT1G24620
|
[EF hand calcium-binding protein family] |
-0.896 |
-0.042 |
| 320 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.892 |
0.078 |
| 321 |
267137_at |
AT2G23410
|
ACPT, cis-prenyltransferase, AtcPT3, CPT, cis-prenyltransferase, cPT3, cis-prenyltransferase 3 |
-0.888 |
0.051 |
| 322 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.880 |
0.048 |
| 323 |
258367_at |
AT3G14370
|
WAG2 |
-0.879 |
0.006 |
| 324 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.877 |
-0.079 |
| 325 |
247781_at |
AT5G58784
|
[Undecaprenyl pyrophosphate synthetase family protein] |
-0.874 |
-0.050 |
| 326 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.871 |
-0.059 |
| 327 |
256283_at |
AT3G12540
|
unknown |
-0.869 |
0.054 |
| 328 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.866 |
-0.078 |
| 329 |
262138_at |
AT1G52660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.863 |
0.002 |
| 330 |
264643_at |
AT1G08990
|
GUX5, Glucuronic Acid Substitution of Xylan 5, PGSIP5, plant glycogenin-like starch initiation protein 5 |
-0.853 |
0.026 |
| 331 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.848 |
0.067 |
| 332 |
253179_at |
AT4G35200
|
unknown |
-0.840 |
0.026 |
| 333 |
264253_at |
AT1G09170
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.835 |
0.042 |
| 334 |
267439_at |
AT2G19060
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.834 |
-0.062 |
| 335 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.834 |
0.031 |
| 336 |
246534_at |
AT5G15890
|
TBL21, TRICHOME BIREFRINGENCE-LIKE 21 |
-0.833 |
0.055 |
| 337 |
251044_at |
AT5G02350
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.831 |
-0.013 |
| 338 |
252582_at |
AT3G45530
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.830 |
-0.004 |
| 339 |
249768_at |
AT5G24100
|
[Leucine-rich repeat protein kinase family protein] |
-0.829 |
0.008 |
| 340 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.825 |
0.001 |
| 341 |
264347_at |
AT1G12040
|
LRX1, leucine-rich repeat/extensin 1 |
-0.824 |
0.013 |
| 342 |
265155_at |
AT1G30990
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.820 |
-0.047 |
| 343 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.817 |
-0.020 |
| 344 |
257418_at |
AT1G30850
|
RSH4, root hair specific 4 |
-0.817 |
0.027 |
| 345 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.813 |
-0.038 |
| 346 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
-0.812 |
0.031 |
| 347 |
260992_at |
AT1G12150
|
unknown |
-0.812 |
0.068 |
| 348 |
252341_at |
AT3G48940
|
[Remorin family protein] |
-0.808 |
-0.028 |
| 349 |
264282_at |
AT1G61840
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.805 |
0.045 |
| 350 |
249185_at |
AT5G43030
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.801 |
0.011 |
| 351 |
267217_at |
AT2G02610
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.798 |
0.008 |
| 352 |
265683_at |
AT2G24400
|
SAUR38, SMALL AUXIN UPREGULATED RNA 38 |
-0.796 |
-0.056 |
| 353 |
259964_at |
AT1G53680
|
ATGSTU28, glutathione S-transferase TAU 28, GSTU28, glutathione S-transferase TAU 28 |
-0.796 |
-0.007 |
| 354 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.795 |
-0.066 |
| 355 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.793 |
-0.040 |
| 356 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
-0.792 |
-0.035 |
| 357 |
258541_at |
AT3G07000
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.785 |
-0.039 |
| 358 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.782 |
0.019 |
| 359 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.777 |
0.069 |
| 360 |
255022_at |
AT4G10350
|
ANAC070, NAC domain containing protein 70, BRN2, BEARSKIN 2, NAC070, NAC domain containing protein 70 |
-0.775 |
0.064 |
| 361 |
266784_at |
AT2G28960
|
[Leucine-rich repeat protein kinase family protein] |
-0.772 |
-0.038 |
| 362 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
-0.768 |
0.025 |
| 363 |
256131_at |
AT1G13600
|
AtbZIP58, basic leucine-zipper 58, bZIP58, basic leucine-zipper 58 |
-0.766 |
0.026 |
| 364 |
259260_at |
AT3G11370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.766 |
-0.051 |
| 365 |
253515_at |
AT4G31320
|
SAUR37, SMALL AUXIN UPREGULATED RNA 37 |
-0.765 |
-0.016 |
| 366 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.765 |
-0.013 |
| 367 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
-0.761 |
0.017 |
| 368 |
258832_at |
AT3G07070
|
[Protein kinase superfamily protein] |
-0.760 |
0.052 |
| 369 |
266011_at |
AT2G37440
|
[DNAse I-like superfamily protein] |
-0.760 |
-0.014 |
| 370 |
247531_at |
AT5G61550
|
[U-box domain-containing protein kinase family protein] |
-0.756 |
0.009 |
| 371 |
257720_at |
AT3G18450
|
[PLAC8 family protein] |
-0.756 |
-0.117 |
| 372 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
-0.756 |
0.124 |
| 373 |
258648_at |
AT3G07900
|
[O-fucosyltransferase family protein] |
-0.755 |
0.000 |
| 374 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.750 |
0.035 |
| 375 |
248212_at |
AT5G54020
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.747 |
0.016 |
| 376 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-0.739 |
0.031 |
| 377 |
266308_at |
AT2G27010
|
CYP705A9, cytochrome P450, family 705, subfamily A, polypeptide 9 |
-0.738 |
0.032 |
| 378 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.737 |
0.022 |
| 379 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.737 |
0.038 |
| 380 |
249868_at |
AT5G23030
|
TET12, tetraspanin12 |
-0.736 |
-0.029 |
| 381 |
262904_at |
AT1G59725
|
[DNAJ heat shock family protein] |
-0.734 |
-0.034 |
| 382 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
-0.733 |
0.120 |
| 383 |
260344_at |
AT1G69240
|
ATMES15, ARABIDOPSIS THALIANA METHYL ESTERASE 15, MES15, methyl esterase 15, RHS9, ROOT HAIR SPECIFIC 9 |
-0.730 |
0.090 |
| 384 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.730 |
0.010 |
| 385 |
264143_at |
AT1G79330
|
AMC6, ATMC5, metacaspase 5, ATMCP2b, metacaspase 2b, MC5, metacaspase 5, MCP2b, metacaspase 2b |
-0.728 |
0.013 |
| 386 |
247475_at |
AT5G62310
|
IRE, INCOMPLETE ROOT HAIR ELONGATION |
-0.726 |
0.057 |
| 387 |
251156_at |
AT3G63060
|
EDL3, EID1-like 3 |
-0.726 |
0.035 |
| 388 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.725 |
0.082 |
| 389 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.723 |
-0.009 |
| 390 |
257924_at |
AT3G23190
|
[HR-like lesion-inducing protein-related] |
-0.723 |
-0.028 |
| 391 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.722 |
0.076 |
| 392 |
249756_at |
AT5G24313
|
unknown |
-0.718 |
0.038 |
| 393 |
255757_at |
AT4G00460
|
ATROPGEF3, ROPGEF3, ROP (rho of plants) guanine nucleotide exchange factor 3 |
-0.715 |
0.037 |
| 394 |
254749_at |
AT4G13130
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.714 |
0.013 |
| 395 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
-0.712 |
-0.077 |
| 396 |
254534_at |
AT4G19680
|
ATIRT2, IRON REGULATED TRANSPORTER 2, IRT2, iron regulated transporter 2 |
-0.711 |
0.085 |
| 397 |
246844_at |
AT5G26660
|
ATMYB86, myb domain protein 86, MYB86, myb domain protein 86 |
-0.706 |
-0.014 |
| 398 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
-0.704 |
-0.074 |
| 399 |
249166_at |
AT5G42840
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.703 |
0.027 |
| 400 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.703 |
0.034 |
| 401 |
267216_at |
AT2G02620
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.703 |
0.000 |
| 402 |
265124_at |
AT1G55430
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.702 |
0.001 |
| 403 |
257595_at |
AT3G24750
|
unknown |
-0.698 |
-0.004 |
| 404 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.698 |
-0.030 |
| 405 |
247674_at |
AT5G59930
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.698 |
-0.021 |
| 406 |
267622_at |
AT2G39690
|
unknown |
-0.698 |
0.002 |
| 407 |
263069_at |
AT2G17590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.693 |
-0.017 |
| 408 |
265329_at |
AT2G18450
|
SDH1-2, succinate dehydrogenase 1-2 |
-0.691 |
-0.001 |
| 409 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.690 |
0.008 |
| 410 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
-0.688 |
0.058 |
| 411 |
248706_at |
AT5G48530
|
unknown |
-0.688 |
0.016 |
| 412 |
246952_at |
AT5G04820
|
ATOFP13, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 13, OFP13, ovate family protein 13 |
-0.688 |
-0.037 |
| 413 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.688 |
0.035 |
| 414 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
-0.679 |
-0.012 |
| 415 |
260752_at |
AT1G49030
|
[PLAC8 family protein] |
-0.677 |
-0.118 |
| 416 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.676 |
0.016 |
| 417 |
253734_at |
AT4G29180
|
RHS16, root hair specific 16 |
-0.671 |
0.004 |
| 418 |
248652_at |
AT5G49270
|
COBL9, COBRA-LIKE 9, DER9, DEFORMED ROOT HAIRS 9, MRH4, MUTANT ROOT HAIR 4, SHV2, SHAVEN 2 |
-0.668 |
0.032 |
| 419 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
-0.667 |
-0.033 |
| 420 |
254774_at |
AT4G13440
|
[Calcium-binding EF-hand family protein] |
-0.667 |
-0.071 |
| 421 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
-0.666 |
0.068 |
| 422 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
-0.666 |
0.017 |
| 423 |
249779_at |
AT5G24230
|
[Lipase class 3-related protein] |
-0.665 |
-0.049 |
| 424 |
253150_at |
AT4G35660
|
unknown |
-0.662 |
0.008 |
| 425 |
256208_at |
AT1G50930
|
unknown |
-0.660 |
0.040 |
| 426 |
264323_at |
AT1G04180
|
YUC9, YUCCA 9 |
-0.660 |
0.016 |
| 427 |
258540_at |
AT3G06990
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.659 |
-0.060 |
| 428 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
-0.659 |
0.077 |
| 429 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.656 |
0.089 |
| 430 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.655 |
0.064 |
| 431 |
257089_at |
AT3G20520
|
GDPDL5, Glycerophosphodiester phosphodiesterase (GDPD) like 5, SVL3, SHV3-like 3 |
-0.655 |
-0.003 |
| 432 |
263542_at |
AT2G21630
|
[Sec23/Sec24 protein transport family protein] |
-0.655 |
-0.032 |
| 433 |
267582_at |
AT2G41970
|
[Protein kinase superfamily protein] |
-0.654 |
0.021 |
| 434 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.653 |
0.118 |
| 435 |
267013_at |
AT2G39180
|
ATCRR2, CCR2, CRINKLY4 related 2 |
-0.649 |
-0.064 |
| 436 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
-0.649 |
-0.030 |
| 437 |
262162_at |
AT1G78020
|
unknown |
-0.649 |
-0.064 |
| 438 |
256359_at |
AT1G66460
|
[Protein kinase superfamily protein] |
-0.648 |
-0.015 |
| 439 |
261514_at |
AT1G71870
|
[MATE efflux family protein] |
-0.644 |
0.012 |
| 440 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.644 |
0.018 |
| 441 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.641 |
-0.060 |
| 442 |
247905_at |
AT5G57400
|
unknown |
-0.641 |
0.051 |
| 443 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
-0.640 |
-0.082 |
| 444 |
247581_at |
AT5G61350
|
[Protein kinase superfamily protein] |
-0.639 |
0.019 |
| 445 |
248715_at |
AT5G48290
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.638 |
-0.033 |
| 446 |
251368_at |
AT3G61380
|
TRM14, TON1 Recruiting Motif 14 |
-0.638 |
-0.024 |
| 447 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.636 |
0.080 |
| 448 |
250778_at |
AT5G05500
|
MOP10 |
-0.636 |
0.038 |
| 449 |
257613_at |
AT3G26610
|
[Pectin lyase-like superfamily protein] |
-0.635 |
0.014 |
| 450 |
254294_at |
AT4G23070
|
ATRBL7, RHOMBOID-like protein 7, RBL7, RHOMBOID-like protein 7 |
-0.634 |
-0.049 |
| 451 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
-0.634 |
0.142 |
| 452 |
263828_at |
AT2G40250
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.634 |
0.041 |
| 453 |
262743_at |
AT1G29020
|
[Calcium-binding EF-hand family protein] |
-0.633 |
-0.045 |
| 454 |
260903_at |
AT1G02460
|
[Pectin lyase-like superfamily protein] |
-0.631 |
0.010 |
| 455 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.631 |
-0.019 |
| 456 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.630 |
0.030 |
| 457 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.630 |
0.020 |
| 458 |
264459_at |
AT1G10160
|
[non-LTR retrotransposon family (LINE), has a 3.9e-39 P-value blast match to GB:AAA67727 reverse transcriptase (LINE-element) (Mus musculus)] |
-0.629 |
-0.039 |
| 459 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-0.628 |
-0.058 |
| 460 |
267037_at |
AT2G38320
|
TBL34, TRICHOME BIREFRINGENCE-LIKE 34 |
-0.627 |
0.005 |
| 461 |
267089_at |
AT2G38300
|
[myb-like HTH transcriptional regulator family protein] |
-0.625 |
0.013 |
| 462 |
249018_at |
AT5G44770
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.624 |
-0.024 |
| 463 |
259576_at |
AT1G35330
|
[RING/U-box superfamily protein] |
-0.623 |
-0.015 |
| 464 |
258017_at |
AT3G19370
|
unknown |
-0.622 |
0.024 |
| 465 |
264495_at |
AT1G27380
|
RIC2, ROP-interactive CRIB motif-containing protein 2 |
-0.618 |
-0.045 |
| 466 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
-0.618 |
0.017 |
| 467 |
254431_at |
AT4G20840
|
[FAD-binding Berberine family protein] |
-0.617 |
-0.033 |
| 468 |
253353_at |
AT4G33730
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.616 |
0.047 |
| 469 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.615 |
-0.032 |
| 470 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
-0.615 |
-0.098 |
| 471 |
254097_at |
AT4G25160
|
[U-box domain-containing protein kinase family protein] |
-0.612 |
-0.034 |
| 472 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.610 |
0.048 |
| 473 |
250204_at |
AT5G13990
|
ATEXO70C2, exocyst subunit exo70 family protein C2, EXO70C2, exocyst subunit exo70 family protein C2 |
-0.610 |
0.013 |
| 474 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
-0.609 |
0.050 |
| 475 |
261777_at |
AT1G76210
|
unknown |
-0.609 |
-0.037 |
| 476 |
256535_at |
AT1G33280
|
ANAC015, NAC domain containing protein 15, BRN1, BEARSKIN 1, NAC015, NAC domain containing protein 15 |
-0.608 |
0.003 |
| 477 |
263458_at |
AT2G22290
|
ATRAB-H1D, ARABIDOPSIS RAB GTPASE HOMOLOG H1D, ATRAB6, ARABIDOPSIS RAB GTPASE HOMOLOG 6, ATRABH1D, RAB GTPase homolog H1D, RAB-H1D, RAB GTPASE HOMOLOG H1D, RABH1d, RAB GTPase homolog H1D |
-0.608 |
0.045 |
| 478 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.608 |
-0.001 |
| 479 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.608 |
-0.013 |
| 480 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.606 |
-0.037 |
| 481 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
-0.605 |
-0.014 |
| 482 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.603 |
-0.020 |
| 483 |
263596_at |
AT2G01900
|
[DNAse I-like superfamily protein] |
-0.600 |
0.003 |
| 484 |
259961_at |
AT1G53700
|
PK3AT, PROTEIN KINASE 3 ARABIDOPSIS THALIANA, WAG1, WAG 1 |
-0.599 |
-0.007 |
| 485 |
252244_at |
AT3G50130
|
unknown |
-0.597 |
-0.029 |
| 486 |
250697_at |
AT5G06800
|
[myb-like HTH transcriptional regulator family protein] |
-0.596 |
-0.039 |
| 487 |
249034_at |
AT5G44160
|
AtIDD8, IDD8, INDETERMINATE DOMAIN 8, NUC, nutcracker |
-0.595 |
0.036 |
| 488 |
262951_at |
AT1G75500
|
UMAMIT5, Usually multiple acids move in and out Transporters 5, WAT1, Walls Are Thin 1 |
-0.595 |
-0.016 |
| 489 |
258035_at |
AT3G21180
|
ACA9, autoinhibited Ca(2+)-ATPase 9, ATACA9 |
-0.595 |
0.046 |
| 490 |
264144_at |
AT1G79320
|
AtMC6, metacaspase 6, AtMCP2c, metacaspase 2c, MC6, metacaspase 6, MCP2c, metacaspase 2c |
-0.590 |
0.082 |
| 491 |
256935_at |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.589 |
-0.033 |
| 492 |
266247_at |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.588 |
0.000 |
| 493 |
247536_at |
AT5G61650
|
CYCP4, CYCP4;2, CYCLIN P4;2 |
-0.587 |
0.060 |
| 494 |
259853_at |
AT1G72300
|
PSY1R, PSY1 receptor |
-0.586 |
-0.007 |
| 495 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
-0.585 |
-0.001 |
| 496 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.585 |
-0.001 |
| 497 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.583 |
-0.051 |
| 498 |
262793_at |
AT1G13110
|
CYP71B7, cytochrome P450, family 71 subfamily B, polypeptide 7 |
-0.583 |
-0.040 |
| 499 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
-0.582 |
-0.034 |
| 500 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.582 |
-0.011 |
| 501 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
-0.580 |
-0.027 |
| 502 |
257117_at |
AT3G20160
|
[Terpenoid synthases superfamily protein] |
-0.580 |
-0.045 |
| 503 |
248218_at |
AT5G53710
|
unknown |
-0.580 |
-0.025 |
| 504 |
262170_at |
AT1G74940
|
unknown |
-0.580 |
0.075 |
| 505 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.580 |
0.049 |
| 506 |
253595_at |
AT4G30830
|
unknown |
-0.579 |
-0.001 |
| 507 |
245979_at |
AT5G13150
|
ATEXO70C1, exocyst subunit exo70 family protein C1, EXO70C1, exocyst subunit exo70 family protein C1 |
-0.576 |
-0.015 |
| 508 |
249186_at |
AT5G43040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.574 |
-0.032 |
| 509 |
260495_at |
AT2G41810
|
unknown |
-0.574 |
-0.019 |
| 510 |
265710_at |
AT2G03370
|
[Glycosyltransferase family 61 protein] |
-0.573 |
0.021 |
| 511 |
248306_at |
AT5G52830
|
ATWRKY27, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 27, WRKY27, WRKY DNA-binding protein 27 |
-0.573 |
0.031 |
| 512 |
253931_at |
AT4G26770
|
[Phosphatidate cytidylyltransferase family protein] |
-0.572 |
0.060 |
| 513 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
-0.571 |
0.110 |
| 514 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.571 |
-0.000 |
| 515 |
258667_at |
AT3G08750
|
[F-box and associated interaction domains-containing protein] |
-0.571 |
-0.035 |
| 516 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.571 |
-0.021 |
| 517 |
261252_at |
AT1G05810
|
ARA, ARA-1, ATRAB11D, ATRABA5E, ARABIDOPSIS THALIANA RAB GTPASE HOMOLOG A5E, RABA5E, RAB GTPase homolog A5E |
-0.570 |
-0.039 |
| 518 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
-0.567 |
0.026 |
| 519 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.567 |
0.090 |
| 520 |
247778_at |
AT5G58750
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.566 |
0.020 |
| 521 |
255028_at |
AT4G09890
|
unknown |
-0.566 |
0.039 |
| 522 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.565 |
-0.050 |
| 523 |
248931_at |
AT5G46040
|
[Major facilitator superfamily protein] |
-0.564 |
-0.075 |
| 524 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
-0.562 |
0.013 |
| 525 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
-0.561 |
0.015 |
| 526 |
245568_at |
AT4G14650
|
unknown |
-0.560 |
0.020 |
| 527 |
256162_at |
AT1G55390
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.560 |
-0.019 |
| 528 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
-0.559 |
-0.005 |
| 529 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.557 |
-0.035 |
| 530 |
252302_at |
AT3G49190
|
[O-acyltransferase (WSD1-like) family protein] |
-0.557 |
0.014 |
| 531 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.556 |
-0.038 |
| 532 |
251364_at |
AT3G61300
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.552 |
-0.001 |
| 533 |
266546_at |
AT2G35270
|
AHL21, AT-hook motif nuclear localized protein 21, GIK, GIANT KILLER |
-0.551 |
-0.040 |
| 534 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
-0.551 |
-0.018 |
| 535 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.551 |
0.013 |
| 536 |
261088_at |
AT1G07590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.549 |
0.002 |
| 537 |
254092_at |
AT4G25090
|
[Riboflavin synthase-like superfamily protein] |
-0.548 |
-0.023 |
| 538 |
257502_at |
AT1G78110
|
unknown |
-0.548 |
-0.059 |
| 539 |
255432_at |
AT4G03330
|
ATSYP123, SYP123, syntaxin of plants 123 |
-0.546 |
-0.022 |
| 540 |
262396_at |
AT1G49470
|
unknown |
-0.543 |
0.034 |
| 541 |
252494_at |
AT3G46760
|
LecRK-S.3, L-type lectin receptor kinase S.3 |
-0.542 |
0.035 |
| 542 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
-0.542 |
0.005 |
| 543 |
255266_at |
AT4G05200
|
CRK25, cysteine-rich RLK (RECEPTOR-like protein kinase) 25 |
-0.541 |
-0.010 |
| 544 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.540 |
-0.011 |
| 545 |
247225_at |
AT5G65090
|
BST1, BRISTLED 1, DER4, DEFORMED ROOT HAIRS 4, MRH3 |
-0.539 |
0.032 |
| 546 |
266069_at |
AT2G18650
|
MEE16, maternal effect embryo arrest 16 |
-0.536 |
-0.062 |
| 547 |
246912_at |
AT5G25820
|
[Exostosin family protein] |
-0.536 |
0.068 |
| 548 |
246802_at |
AT5G27000
|
ATK4, kinesin 4, KATD, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA D |
-0.535 |
-0.040 |
| 549 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.535 |
0.065 |
| 550 |
249367_at |
AT5G40630
|
[Ubiquitin-like superfamily protein] |
-0.534 |
0.019 |
| 551 |
267427_at |
AT2G34830
|
AtWRKY35, WRKY DNA-BINDING PROTEIN 35, MEE24, maternal effect embryo arrest 24, WRKY35, WRKY DNA-binding protein 35 |
-0.534 |
-0.029 |
| 552 |
252964_at |
AT4G38830
|
CRK26, cysteine-rich RLK (RECEPTOR-like protein kinase) 26 |
-0.534 |
-0.030 |
| 553 |
267409_at |
AT2G34910
|
unknown |
-0.533 |
0.013 |
| 554 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.530 |
-0.051 |
| 555 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
-0.529 |
-0.038 |
| 556 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.528 |
-0.027 |
| 557 |
255175_at |
AT4G07960
|
ATCSLC12, Cellulose-synthase-like C12, CSLC12, CELLULOSE-SYNTHASE LIKE C12, CSLC12, Cellulose-synthase-like C12 |
-0.527 |
-0.039 |
| 558 |
266369_at |
AT2G41370
|
BOP2, BLADE ON PETIOLE2 |
-0.527 |
-0.086 |
| 559 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.527 |
-0.021 |
| 560 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
-0.526 |
0.028 |
| 561 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
-0.523 |
-0.055 |
| 562 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
-0.522 |
-0.004 |
| 563 |
267222_at |
AT2G43880
|
[Pectin lyase-like superfamily protein] |
-0.521 |
-0.010 |
| 564 |
254284_at |
AT4G22910
|
CCS52A1, cell cycle switch protein 52 A1, FZR2, FIZZY-related 2 |
-0.520 |
-0.011 |
| 565 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.520 |
-0.027 |
| 566 |
257371_at |
AT2G47810
|
NF-YB5, nuclear factor Y, subunit B5 |
-0.519 |
0.063 |
| 567 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.519 |
-0.033 |
| 568 |
246807_at |
AT5G27100
|
ATGLR2.1, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 2.1, GLR2.1, glutamate receptor 2.1 |
-0.519 |
0.003 |
| 569 |
249790_at |
AT5G24290
|
MEB2, MEMBRANE OF ER BODY 2 |
-0.518 |
0.004 |
| 570 |
253910_at |
AT4G27290
|
[S-locus lectin protein kinase family protein] |
-0.518 |
-0.042 |
| 571 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
-0.518 |
-0.030 |
| 572 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.518 |
0.034 |
| 573 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
-0.518 |
0.060 |
| 574 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.518 |
-0.051 |
| 575 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.518 |
-0.022 |
| 576 |
245574_at |
AT4G14750
|
IQD19, IQ-domain 19 |
-0.518 |
0.014 |
| 577 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.517 |
-0.005 |
| 578 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
-0.517 |
-0.140 |
| 579 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.516 |
-0.041 |
| 580 |
245300_at |
AT4G16350
|
CBL6, calcineurin B-like protein 6, SCABP2, SOS3-LIKE CALCIUM BINDING PROTEIN 2 |
-0.516 |
-0.015 |
| 581 |
263883_at |
AT2G21830
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.516 |
-0.019 |
| 582 |
254173_at |
AT4G24580
|
REN1, ROP1 ENHANCER 1 |
-0.515 |
0.075 |
| 583 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.514 |
0.008 |
| 584 |
262620_at |
AT1G06540
|
unknown |
-0.514 |
-0.021 |
| 585 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.514 |
0.011 |
| 586 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.512 |
0.015 |
| 587 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.512 |
-0.028 |
| 588 |
265250_at |
AT2G01950
|
BRL2, BRI1-like 2, VH1, VASCULAR HIGHWAY 1 |
-0.512 |
-0.023 |
| 589 |
264988_at |
AT1G27140
|
ATGSTU14, glutathione S-transferase tau 14, GST13, GLUTATHIONE S-TRANSFERASE 13, GSTU14, glutathione S-transferase tau 14 |
-0.512 |
0.037 |
| 590 |
246826_at |
AT5G26310
|
UGT72E3 |
-0.511 |
0.004 |
| 591 |
254912_at |
AT4G11230
|
[Riboflavin synthase-like superfamily protein] |
-0.511 |
-0.064 |
| 592 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
-0.509 |
-0.001 |
| 593 |
256778_at |
AT3G13782
|
NAP1;4, nucleosome assembly protein1;4, NFA04, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 04, NFA4, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 4 |
-0.509 |
0.067 |
| 594 |
254240_at |
AT4G23496
|
SP1L5, SPIRAL1-like5 |
-0.507 |
0.004 |
| 595 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
-0.506 |
-0.038 |
| 596 |
255074_at |
AT4G09100
|
[RING/U-box superfamily protein] |
-0.506 |
-0.028 |
| 597 |
257277_at |
AT3G14470
|
[NB-ARC domain-containing disease resistance protein] |
-0.506 |
0.048 |
| 598 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.506 |
0.010 |
| 599 |
262664_at |
AT1G13970
|
unknown |
-0.506 |
0.033 |
| 600 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.505 |
0.044 |