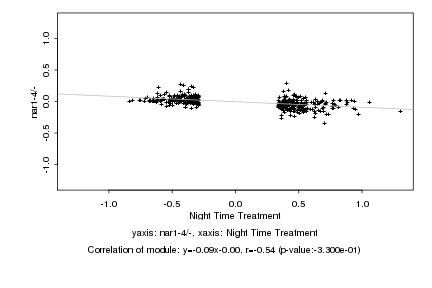
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Night Time Treatment |
Log2 signal ratio
nar1-4/- |
| 1 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
1.301 |
-0.146 |
| 2 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
1.053 |
-0.014 |
| 3 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.971 |
-0.198 |
| 4 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.947 |
-0.119 |
| 5 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
0.933 |
0.016 |
| 6 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.925 |
-0.098 |
| 7 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.915 |
0.032 |
| 8 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.883 |
-0.003 |
| 9 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
0.876 |
-0.045 |
| 10 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.870 |
0.030 |
| 11 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.825 |
0.023 |
| 12 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.816 |
0.019 |
| 13 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.811 |
-0.085 |
| 14 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.786 |
-0.061 |
| 15 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
0.779 |
-0.036 |
| 16 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.773 |
0.018 |
| 17 |
252661_at |
AT3G44450
|
unknown |
0.767 |
-0.096 |
| 18 |
261247_at |
AT1G20070
|
unknown |
0.764 |
-0.097 |
| 19 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
0.760 |
-0.027 |
| 20 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.759 |
-0.077 |
| 21 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.731 |
-0.200 |
| 22 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.713 |
-0.192 |
| 23 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.712 |
-0.010 |
| 24 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.711 |
-0.045 |
| 25 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.709 |
0.128 |
| 26 |
265892_at |
AT2G15020
|
unknown |
0.704 |
-0.021 |
| 27 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.698 |
-0.344 |
| 28 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.691 |
-0.147 |
| 29 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.689 |
-0.079 |
| 30 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.688 |
-0.153 |
| 31 |
260956_at |
AT1G06040
|
BBX24, B-box domain protein 24, STO, SALT TOLERANCE |
0.684 |
0.012 |
| 32 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
0.683 |
0.030 |
| 33 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.680 |
-0.107 |
| 34 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.669 |
-0.152 |
| 35 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.669 |
-0.012 |
| 36 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.662 |
-0.025 |
| 37 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.654 |
-0.142 |
| 38 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.644 |
0.014 |
| 39 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
0.641 |
-0.039 |
| 40 |
257057_at |
AT3G15310
|
unknown |
0.631 |
-0.035 |
| 41 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
0.630 |
-0.184 |
| 42 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.626 |
0.044 |
| 43 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
0.625 |
-0.091 |
| 44 |
256603_at |
AT3G28270
|
unknown |
0.624 |
-0.245 |
| 45 |
252010_at |
AT3G52740
|
unknown |
0.623 |
-0.034 |
| 46 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.623 |
-0.143 |
| 47 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
0.616 |
-0.015 |
| 48 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.613 |
-0.003 |
| 49 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.603 |
-0.126 |
| 50 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.600 |
-0.007 |
| 51 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.589 |
-0.114 |
| 52 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.568 |
-0.109 |
| 53 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
0.560 |
-0.119 |
| 54 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.556 |
-0.033 |
| 55 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.556 |
-0.006 |
| 56 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
0.555 |
0.018 |
| 57 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.554 |
-0.144 |
| 58 |
251058_at |
AT5G01790
|
unknown |
0.554 |
-0.001 |
| 59 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
0.552 |
-0.022 |
| 60 |
256518_at |
AT1G66080
|
unknown |
0.550 |
0.069 |
| 61 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.549 |
-0.123 |
| 62 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
0.546 |
-0.128 |
| 63 |
251725_at |
AT3G56260
|
unknown |
0.543 |
-0.027 |
| 64 |
249134_at |
AT5G43150
|
unknown |
0.542 |
-0.054 |
| 65 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.541 |
-0.033 |
| 66 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
0.540 |
-0.053 |
| 67 |
246125_at |
AT5G19875
|
unknown |
0.540 |
-0.061 |
| 68 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.537 |
-0.170 |
| 69 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.532 |
-0.098 |
| 70 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.531 |
0.048 |
| 71 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.530 |
-0.158 |
| 72 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
0.528 |
-0.042 |
| 73 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
0.527 |
-0.127 |
| 74 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.526 |
-0.104 |
| 75 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
0.520 |
-0.068 |
| 76 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.519 |
-0.027 |
| 77 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
0.517 |
-0.011 |
| 78 |
251811_at |
AT3G54990
|
SMZ, SCHLAFMUTZE |
0.517 |
0.032 |
| 79 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
0.517 |
-0.112 |
| 80 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.515 |
-0.001 |
| 81 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.514 |
-0.044 |
| 82 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.512 |
-0.074 |
| 83 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.512 |
-0.137 |
| 84 |
251292_at |
AT3G61920
|
unknown |
0.511 |
0.086 |
| 85 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.505 |
-0.067 |
| 86 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.504 |
-0.072 |
| 87 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.503 |
-0.178 |
| 88 |
249622_at |
AT5G37550
|
unknown |
0.501 |
-0.067 |
| 89 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.499 |
-0.115 |
| 90 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.494 |
0.079 |
| 91 |
245078_at |
AT2G23340
|
DEAR3, DREB and EAR motif protein 3 |
0.494 |
-0.039 |
| 92 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.492 |
-0.091 |
| 93 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.490 |
-0.012 |
| 94 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
0.487 |
-0.031 |
| 95 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.487 |
-0.141 |
| 96 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.486 |
0.022 |
| 97 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.484 |
-0.159 |
| 98 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.481 |
-0.004 |
| 99 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.479 |
0.021 |
| 100 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.477 |
0.082 |
| 101 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.475 |
-0.010 |
| 102 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.474 |
-0.046 |
| 103 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
0.473 |
-0.049 |
| 104 |
266757_at |
AT2G46940
|
unknown |
0.472 |
0.090 |
| 105 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.468 |
-0.035 |
| 106 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
0.466 |
-0.151 |
| 107 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
0.464 |
-0.048 |
| 108 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.462 |
-0.097 |
| 109 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
0.461 |
-0.002 |
| 110 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.460 |
-0.236 |
| 111 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
0.460 |
-0.068 |
| 112 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
0.460 |
-0.122 |
| 113 |
252427_at |
AT3G47640
|
PYE, POPEYE |
0.459 |
0.112 |
| 114 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
0.459 |
-0.043 |
| 115 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.458 |
-0.144 |
| 116 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
0.457 |
0.035 |
| 117 |
253859_at |
AT4G27657
|
unknown |
0.453 |
-0.026 |
| 118 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
0.453 |
0.106 |
| 119 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.451 |
-0.068 |
| 120 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.450 |
0.015 |
| 121 |
251896_at |
AT3G54390
|
[sequence-specific DNA binding transcription factors] |
0.449 |
-0.027 |
| 122 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.447 |
-0.025 |
| 123 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
0.447 |
0.005 |
| 124 |
257375_at |
AT2G38640
|
unknown |
0.444 |
-0.017 |
| 125 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.442 |
0.009 |
| 126 |
258807_at |
AT3G04030
|
MYR2 |
0.441 |
-0.054 |
| 127 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.440 |
-0.083 |
| 128 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
0.438 |
-0.113 |
| 129 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.437 |
-0.102 |
| 130 |
260455_at |
AT1G72500
|
[LOCATED IN: plasma membrane] |
0.436 |
-0.053 |
| 131 |
259985_at |
AT1G76620
|
unknown |
0.435 |
-0.007 |
| 132 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.435 |
-0.000 |
| 133 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.434 |
-0.212 |
| 134 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.434 |
-0.115 |
| 135 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
0.434 |
0.002 |
| 136 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.433 |
0.021 |
| 137 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
0.433 |
-0.035 |
| 138 |
252888_at |
AT4G39210
|
APL3 |
0.433 |
-0.115 |
| 139 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.432 |
-0.154 |
| 140 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.425 |
-0.121 |
| 141 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
0.425 |
-0.056 |
| 142 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
0.422 |
-0.113 |
| 143 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
0.422 |
0.033 |
| 144 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
0.418 |
-0.044 |
| 145 |
260491_at |
AT1G51440
|
DALL2, DAD1-Like Lipase 2 |
0.418 |
-0.013 |
| 146 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
0.417 |
-0.000 |
| 147 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.417 |
0.176 |
| 148 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
0.416 |
0.023 |
| 149 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.415 |
-0.063 |
| 150 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
0.414 |
-0.053 |
| 151 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
0.412 |
0.007 |
| 152 |
264479_at |
AT1G77280
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.411 |
-0.114 |
| 153 |
263829_at |
AT2G40435
|
unknown |
0.410 |
-0.129 |
| 154 |
251644_at |
AT3G57540
|
[Remorin family protein] |
0.410 |
-0.064 |
| 155 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
0.408 |
-0.044 |
| 156 |
263545_at |
AT2G21560
|
unknown |
0.406 |
-0.101 |
| 157 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
0.403 |
-0.003 |
| 158 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.401 |
-0.070 |
| 159 |
248205_at |
AT5G54300
|
unknown |
0.400 |
0.087 |
| 160 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.400 |
0.296 |
| 161 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.398 |
-0.060 |
| 162 |
257925_at |
AT3G23170
|
unknown |
0.396 |
-0.002 |
| 163 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
0.396 |
-0.115 |
| 164 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
0.393 |
0.030 |
| 165 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.393 |
-0.041 |
| 166 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
0.393 |
-0.041 |
| 167 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
0.393 |
-0.006 |
| 168 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.392 |
0.069 |
| 169 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
0.391 |
-0.007 |
| 170 |
247055_at |
AT5G66740
|
unknown |
0.388 |
-0.082 |
| 171 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
0.386 |
0.094 |
| 172 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
0.386 |
-0.082 |
| 173 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.386 |
-0.040 |
| 174 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.386 |
-0.159 |
| 175 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.385 |
0.012 |
| 176 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.385 |
0.036 |
| 177 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
0.384 |
-0.015 |
| 178 |
264636_at |
AT1G65490
|
unknown |
0.381 |
0.085 |
| 179 |
264583_at |
AT1G05170
|
[Galactosyltransferase family protein] |
0.379 |
-0.059 |
| 180 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
0.379 |
-0.105 |
| 181 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.379 |
-0.130 |
| 182 |
248232_at |
AT5G53760
|
ATMLO11, MILDEW RESISTANCE LOCUS O 11, MLO11, MILDEW RESISTANCE LOCUS O 11 |
0.379 |
-0.006 |
| 183 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
0.378 |
-0.074 |
| 184 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.377 |
-0.040 |
| 185 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
0.377 |
-0.064 |
| 186 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
0.376 |
-0.131 |
| 187 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.376 |
0.162 |
| 188 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
0.376 |
-0.031 |
| 189 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
0.375 |
-0.069 |
| 190 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
0.374 |
0.025 |
| 191 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.373 |
-0.073 |
| 192 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
0.371 |
-0.046 |
| 193 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
0.370 |
0.002 |
| 194 |
245352_at |
AT4G15490
|
UGT84A3 |
0.368 |
-0.008 |
| 195 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
0.368 |
-0.100 |
| 196 |
251839_at |
AT3G54950
|
PLA IIIA, patatin-like protein 6, PLP7, PATATIN-LIKE PROTEIN 7, pPLAIIIbeta, patatin-related phospholipase IIIbeta |
0.365 |
-0.040 |
| 197 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.362 |
-0.049 |
| 198 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.361 |
-0.263 |
| 199 |
253145_at |
AT4G35560
|
DAW1, DUO1-activated WD40 1 |
0.359 |
-0.019 |
| 200 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
0.359 |
-0.212 |
| 201 |
254693_at |
AT4G17880
|
MYC4 |
0.358 |
-0.026 |
| 202 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
0.357 |
-0.071 |
| 203 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
0.356 |
-0.121 |
| 204 |
257916_at |
AT3G23210
|
bHLH34, basic Helix-Loop-Helix 34 |
0.356 |
-0.107 |
| 205 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.355 |
-0.050 |
| 206 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
0.354 |
-0.075 |
| 207 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
0.354 |
-0.041 |
| 208 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
0.353 |
-0.059 |
| 209 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
0.349 |
-0.041 |
| 210 |
264909_at |
AT2G17300
|
unknown |
0.349 |
-0.022 |
| 211 |
258017_at |
AT3G19370
|
unknown |
0.349 |
-0.086 |
| 212 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.348 |
-0.035 |
| 213 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
0.345 |
-0.067 |
| 214 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.345 |
-0.041 |
| 215 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
0.344 |
0.065 |
| 216 |
266299_at |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
0.343 |
-0.003 |
| 217 |
261921_at |
AT1G65900
|
unknown |
0.343 |
-0.093 |
| 218 |
258119_at |
AT3G14720
|
ATMPK19, ARABIDOPSIS THALIANA MAP KINASE 19, MPK19, MAP kinase 19 |
0.342 |
-0.055 |
| 219 |
253219_at |
AT4G34990
|
AtMYB32, myb domain protein 32, MYB32, myb domain protein 32 |
0.341 |
-0.037 |
| 220 |
246001_at |
AT5G20790
|
unknown |
0.340 |
-0.027 |
| 221 |
267538_at |
AT2G41870
|
[Remorin family protein] |
0.340 |
-0.055 |
| 222 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
0.340 |
0.027 |
| 223 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.340 |
-0.067 |
| 224 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
0.340 |
-0.080 |
| 225 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.842 |
0.005 |
| 226 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.819 |
0.031 |
| 227 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.761 |
0.018 |
| 228 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.754 |
0.025 |
| 229 |
265387_at |
AT2G20670
|
unknown |
-0.724 |
0.016 |
| 230 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.712 |
0.053 |
| 231 |
247474_at |
AT5G62280
|
unknown |
-0.700 |
0.079 |
| 232 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.684 |
0.009 |
| 233 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.680 |
0.018 |
| 234 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.679 |
0.009 |
| 235 |
258225_at |
AT3G15630
|
unknown |
-0.674 |
0.020 |
| 236 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.663 |
0.010 |
| 237 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.657 |
0.008 |
| 238 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
-0.654 |
0.049 |
| 239 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.649 |
0.019 |
| 240 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.649 |
0.014 |
| 241 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.642 |
0.005 |
| 242 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.626 |
-0.002 |
| 243 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.621 |
0.023 |
| 244 |
264238_at |
AT1G54740
|
unknown |
-0.619 |
0.026 |
| 245 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.619 |
0.142 |
| 246 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.618 |
0.037 |
| 247 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.617 |
0.073 |
| 248 |
266658_at |
AT2G25735
|
unknown |
-0.609 |
0.238 |
| 249 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.599 |
0.007 |
| 250 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.597 |
0.100 |
| 251 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.589 |
-0.036 |
| 252 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
-0.585 |
0.047 |
| 253 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.584 |
0.034 |
| 254 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
-0.574 |
0.039 |
| 255 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.569 |
0.036 |
| 256 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.566 |
0.045 |
| 257 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-0.566 |
0.124 |
| 258 |
246929_at |
AT5G25210
|
unknown |
-0.551 |
-0.011 |
| 259 |
248509_at |
AT5G50335
|
unknown |
-0.548 |
-0.074 |
| 260 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.546 |
-0.010 |
| 261 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.545 |
0.155 |
| 262 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.531 |
-0.001 |
| 263 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.526 |
0.036 |
| 264 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.525 |
-0.059 |
| 265 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.524 |
0.092 |
| 266 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
-0.523 |
0.103 |
| 267 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
-0.521 |
0.013 |
| 268 |
251373_at |
AT3G60530
|
GATA4, GATA transcription factor 4 |
-0.520 |
0.022 |
| 269 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.520 |
-0.019 |
| 270 |
247878_at |
AT5G57760
|
unknown |
-0.511 |
0.050 |
| 271 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
-0.503 |
-0.051 |
| 272 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
-0.501 |
0.042 |
| 273 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.498 |
0.032 |
| 274 |
267393_at |
AT2G44500
|
[O-fucosyltransferase family protein] |
-0.498 |
0.008 |
| 275 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
-0.491 |
0.048 |
| 276 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
-0.489 |
0.015 |
| 277 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
-0.487 |
0.098 |
| 278 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.482 |
0.092 |
| 279 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.475 |
0.025 |
| 280 |
264379_at |
AT2G25200
|
unknown |
-0.473 |
0.051 |
| 281 |
247754_at |
AT5G59080
|
unknown |
-0.469 |
-0.029 |
| 282 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.463 |
0.108 |
| 283 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.461 |
0.031 |
| 284 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.457 |
-0.019 |
| 285 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
-0.453 |
0.060 |
| 286 |
253802_at |
AT4G28180
|
unknown |
-0.451 |
0.039 |
| 287 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.451 |
0.019 |
| 288 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.446 |
-0.008 |
| 289 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
-0.446 |
0.085 |
| 290 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
-0.446 |
0.071 |
| 291 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.445 |
0.079 |
| 292 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.444 |
0.058 |
| 293 |
265539_at |
AT2G15830
|
unknown |
-0.443 |
0.044 |
| 294 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.438 |
0.160 |
| 295 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
-0.436 |
0.283 |
| 296 |
260799_at |
AT1G78270
|
AtUGT85A4, UDP-glucosyl transferase 85A4, UGT85A4, UDP-glucosyl transferase 85A4 |
-0.433 |
0.088 |
| 297 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
-0.432 |
0.076 |
| 298 |
252573_at |
AT3G45260
|
[C2H2-like zinc finger protein] |
-0.432 |
0.019 |
| 299 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.431 |
0.082 |
| 300 |
256262_at |
AT3G12150
|
unknown |
-0.431 |
0.003 |
| 301 |
246584_at |
AT5G14730
|
unknown |
-0.430 |
0.085 |
| 302 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.428 |
0.014 |
| 303 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.427 |
0.043 |
| 304 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
-0.422 |
0.120 |
| 305 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.421 |
0.110 |
| 306 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.416 |
-0.019 |
| 307 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
-0.411 |
0.265 |
| 308 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.409 |
0.011 |
| 309 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.408 |
0.022 |
| 310 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
-0.407 |
0.031 |
| 311 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
-0.407 |
0.044 |
| 312 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.405 |
0.012 |
| 313 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-0.405 |
0.088 |
| 314 |
245353_at |
AT4G16000
|
unknown |
-0.404 |
0.127 |
| 315 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.402 |
-0.011 |
| 316 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.400 |
-0.083 |
| 317 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.400 |
0.024 |
| 318 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
-0.399 |
-0.006 |
| 319 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.398 |
0.032 |
| 320 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.397 |
-0.086 |
| 321 |
255285_at |
AT4G04630
|
unknown |
-0.397 |
0.018 |
| 322 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.395 |
0.107 |
| 323 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.394 |
-0.037 |
| 324 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
-0.393 |
0.062 |
| 325 |
253322_at |
AT4G33980
|
unknown |
-0.386 |
0.018 |
| 326 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
-0.385 |
-0.007 |
| 327 |
266016_at |
AT2G18670
|
[RING/U-box superfamily protein] |
-0.384 |
0.038 |
| 328 |
247212_at |
AT5G65040
|
unknown |
-0.384 |
0.045 |
| 329 |
251080_at |
AT5G02010
|
ATROPGEF7, ROPGEF7, ROP (rho of plants) guanine nucleotide exchange factor 7 |
-0.380 |
0.036 |
| 330 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
-0.378 |
0.200 |
| 331 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-0.377 |
0.000 |
| 332 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.377 |
0.018 |
| 333 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.376 |
0.202 |
| 334 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
-0.375 |
0.076 |
| 335 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.374 |
0.058 |
| 336 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
-0.374 |
-0.011 |
| 337 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
-0.372 |
0.044 |
| 338 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.372 |
0.158 |
| 339 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.370 |
0.022 |
| 340 |
264445_at |
AT1G27290
|
unknown |
-0.369 |
0.021 |
| 341 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.369 |
0.085 |
| 342 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
-0.368 |
0.029 |
| 343 |
254954_at |
AT4G10910
|
unknown |
-0.368 |
0.082 |
| 344 |
267209_at |
AT2G30930
|
unknown |
-0.366 |
0.046 |
| 345 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
-0.365 |
0.013 |
| 346 |
246506_at |
AT5G16110
|
unknown |
-0.365 |
0.011 |
| 347 |
257894_at |
AT3G17100
|
AIF3, ATBS1 Interacting Factor 3 |
-0.362 |
0.003 |
| 348 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.360 |
-0.013 |
| 349 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.359 |
0.027 |
| 350 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
-0.358 |
0.006 |
| 351 |
267557_at |
AT2G32710
|
ACK2, CYCLIN-DEPENDENT KINASE INHIBITOR 2, ICK7, INTERACTORS OF CDC2 KINASE 7, KRP4, KRP4, KIP-RELATED PROTEIN 4 |
-0.357 |
0.005 |
| 352 |
258704_at |
AT3G09780
|
ATCRR1, CCR1, CRINKLY4 related 1 |
-0.356 |
-0.002 |
| 353 |
266984_at |
AT2G39570
|
ACR9, ACT domain repeats 9 |
-0.356 |
-0.024 |
| 354 |
254506_at |
AT4G20140
|
GSO1, GASSHO1 |
-0.356 |
-0.028 |
| 355 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.355 |
0.071 |
| 356 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
-0.353 |
0.106 |
| 357 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
-0.350 |
-0.098 |
| 358 |
250107_at |
AT5G15330
|
ATSPX4, ARABIDOPSIS THALIANA SPX DOMAIN GENE 4, SPX4, SPX domain gene 4 |
-0.349 |
0.063 |
| 359 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.348 |
0.240 |
| 360 |
264590_at |
AT2G17710
|
unknown |
-0.346 |
0.049 |
| 361 |
264304_at |
AT1G78895
|
[Reticulon family protein] |
-0.346 |
-0.001 |
| 362 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.345 |
0.049 |
| 363 |
250327_at |
AT5G12050
|
unknown |
-0.344 |
0.070 |
| 364 |
256225_at |
AT1G56220
|
[Dormancy/auxin associated family protein] |
-0.343 |
0.022 |
| 365 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
-0.342 |
-0.000 |
| 366 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.341 |
0.075 |
| 367 |
247585_at |
AT5G60680
|
unknown |
-0.341 |
0.040 |
| 368 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.339 |
-0.024 |
| 369 |
260362_at |
AT1G70530
|
CRK3, cysteine-rich RLK (RECEPTOR-like protein kinase) 3 |
-0.338 |
0.018 |
| 370 |
248302_at |
AT5G53160
|
PYL8, PYR1-like 8, RCAR3, regulatory components of ABA receptor 3 |
-0.338 |
0.012 |
| 371 |
248623_at |
AT5G49170
|
unknown |
-0.337 |
0.001 |
| 372 |
266428_at |
AT2G07180
|
[Protein kinase superfamily protein] |
-0.337 |
-0.007 |
| 373 |
265648_at |
AT2G27500
|
[Glycosyl hydrolase superfamily protein] |
-0.336 |
-0.009 |
| 374 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-0.336 |
0.233 |
| 375 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.335 |
-0.047 |
| 376 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
-0.335 |
0.048 |
| 377 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-0.334 |
0.101 |
| 378 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.332 |
0.005 |
| 379 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.332 |
-0.008 |
| 380 |
249872_at |
AT5G23130
|
[Peptidoglycan-binding LysM domain-containing protein] |
-0.332 |
0.008 |
| 381 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.331 |
0.066 |
| 382 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.331 |
-0.022 |
| 383 |
259015_at |
AT3G07350
|
unknown |
-0.330 |
0.041 |
| 384 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
-0.329 |
-0.022 |
| 385 |
261143_at |
AT1G19770
|
ATPUP14, purine permease 14, PUP14, purine permease 14 |
-0.329 |
0.042 |
| 386 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
-0.329 |
0.001 |
| 387 |
247979_at |
AT5G56750
|
NDL1, N-MYC downregulated-like 1 |
-0.329 |
-0.010 |
| 388 |
261252_at |
AT1G05810
|
ARA, ARA-1, ATRAB11D, ATRABA5E, ARABIDOPSIS THALIANA RAB GTPASE HOMOLOG A5E, RABA5E, RAB GTPase homolog A5E |
-0.328 |
0.006 |
| 389 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
-0.327 |
0.038 |
| 390 |
260167_at |
AT1G71970
|
unknown |
-0.327 |
0.037 |
| 391 |
250753_at |
AT5G05860
|
UGT76C2, UDP-glucosyl transferase 76C2 |
-0.326 |
-0.007 |
| 392 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-0.323 |
0.031 |
| 393 |
261402_at |
AT1G79670
|
RFO1, RESISTANCE TO FUSARIUM OXYSPORUM 1, WAKL22 |
-0.322 |
0.007 |
| 394 |
261711_at |
AT1G32700
|
[PLATZ transcription factor family protein] |
-0.321 |
0.003 |
| 395 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.321 |
-0.025 |
| 396 |
250000_at |
AT5G18650
|
MIEL1, MYB30-Interacting E3 Ligase 1 |
-0.320 |
0.012 |
| 397 |
252464_at |
AT3G47160
|
[RING/U-box superfamily protein] |
-0.320 |
0.041 |
| 398 |
258472_at |
AT3G06080
|
TBL10, TRICHOME BIREFRINGENCE-LIKE 10 |
-0.319 |
0.024 |
| 399 |
250217_at |
AT5G14120
|
[Major facilitator superfamily protein] |
-0.319 |
0.032 |
| 400 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.318 |
0.120 |
| 401 |
250877_at |
AT5G04040
|
SDP1, SUGAR-DEPENDENT1 |
-0.318 |
0.078 |
| 402 |
254424_at |
AT4G21510
|
AtFBS2, FBS2, F-box stress induced 2 |
-0.318 |
0.014 |
| 403 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
-0.318 |
0.050 |
| 404 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
-0.317 |
0.067 |
| 405 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
-0.316 |
0.062 |
| 406 |
257990_at |
AT3G19860
|
bHLH121, basic Helix-Loop-Helix 121 |
-0.316 |
0.021 |
| 407 |
258468_at |
AT3G06070
|
unknown |
-0.315 |
-0.022 |
| 408 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
-0.315 |
0.053 |
| 409 |
264512_at |
AT1G09575
|
unknown |
-0.313 |
0.004 |
| 410 |
258262_at |
AT3G15770
|
unknown |
-0.313 |
-0.010 |
| 411 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
-0.313 |
0.087 |
| 412 |
250524_at |
AT5G08520
|
[Duplicated homeodomain-like superfamily protein] |
-0.310 |
0.053 |
| 413 |
252040_at |
AT3G52060
|
AtGnTL, GnTL, beta-1,6-N-acetylglucosaminyl transferase-like |
-0.310 |
0.010 |
| 414 |
263218_at |
AT1G30570
|
HERK2, hercules receptor kinase 2 |
-0.310 |
0.031 |
| 415 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.310 |
-0.079 |
| 416 |
260317_at |
AT1G63800
|
UBC5, ubiquitin-conjugating enzyme 5 |
-0.308 |
0.047 |
| 417 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.308 |
0.051 |
| 418 |
259164_at |
AT3G01770
|
ATBET10, bromodomain and extraterminal domain protein 10, BET10, bromodomain and extraterminal domain protein 10 |
-0.308 |
-0.006 |
| 419 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
-0.308 |
-0.010 |
| 420 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.307 |
0.067 |
| 421 |
250008_at |
AT5G18630
|
[alpha/beta-Hydrolases superfamily protein] |
-0.306 |
0.056 |
| 422 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.305 |
-0.005 |
| 423 |
249345_at |
AT5G40740
|
AUG6, augmin subunit 6 |
-0.305 |
-0.006 |
| 424 |
265451_at |
AT2G46490
|
unknown |
-0.305 |
-0.012 |
| 425 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
-0.305 |
0.030 |
| 426 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
-0.304 |
0.043 |
| 427 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
-0.304 |
0.088 |
| 428 |
248901_at |
AT5G46410
|
SSP4, SCP1-like small phosphatase 4 |
-0.304 |
-0.042 |
| 429 |
250124_at |
AT5G16480
|
AtPFA-DSP5, PFA-DSP5, plant and fungi atypical dual-specificity phosphatase 5 |
-0.303 |
-0.024 |
| 430 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
-0.302 |
0.047 |
| 431 |
264841_at |
AT1G03740
|
[Protein kinase superfamily protein] |
-0.301 |
0.027 |
| 432 |
261253_at |
AT1G05840
|
[Eukaryotic aspartyl protease family protein] |
-0.301 |
0.008 |
| 433 |
260303_at |
AT1G70520
|
ASG6, ALTERED SEED GERMINATION 6, CRK2, cysteine-rich RLK (RECEPTOR-like protein kinase) 2 |
-0.300 |
0.030 |
| 434 |
250692_at |
AT5G06560
|
unknown |
-0.300 |
0.017 |
| 435 |
264889_at |
AT1G23050
|
hydroxyproline-rich glycoprotein family protein |
-0.299 |
0.039 |
| 436 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.299 |
0.069 |
| 437 |
249824_at |
AT5G23380
|
unknown |
-0.299 |
-0.009 |
| 438 |
265481_at |
AT2G15960
|
unknown |
-0.299 |
0.021 |
| 439 |
252765_at |
AT3G42800
|
unknown |
-0.298 |
-0.050 |
| 440 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.297 |
0.088 |
| 441 |
258647_at |
AT3G07870
|
[F-box and associated interaction domains-containing protein] |
-0.296 |
-0.052 |
| 442 |
250132_at |
AT5G16560
|
KAN, KANADI, KAN1, KANADI 1 |
-0.295 |
-0.030 |
| 443 |
246755_at |
AT5G27920
|
[F-box family protein] |
-0.295 |
-0.033 |
| 444 |
251050_at |
AT5G02440
|
unknown |
-0.295 |
0.108 |
| 445 |
265266_at |
AT2G42890
|
AML2, MEI2-like 2, ML2, MEI2-like 2 |
-0.295 |
-0.032 |
| 446 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
-0.293 |
-0.008 |
| 447 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.292 |
0.093 |