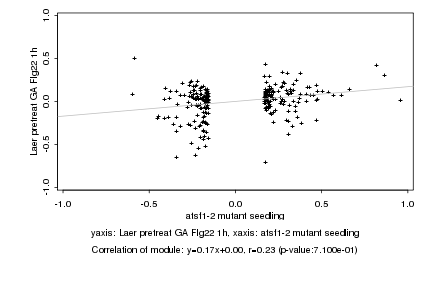
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
atsf1-2 mutant seedling |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
262719_at |
AT1G43590
|
unknown |
0.955 |
0.021 |
| 2 |
255811_at |
AT4G10250
|
ATHSP22.0 |
0.860 |
0.311 |
| 3 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
0.815 |
0.430 |
| 4 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.657 |
0.150 |
| 5 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
0.614 |
0.074 |
| 6 |
263515_at |
AT2G21640
|
unknown |
0.565 |
0.077 |
| 7 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.536 |
0.110 |
| 8 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
0.503 |
0.123 |
| 9 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.474 |
0.032 |
| 10 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.474 |
0.120 |
| 11 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
0.469 |
0.012 |
| 12 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.467 |
-0.218 |
| 13 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.466 |
0.197 |
| 14 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.451 |
0.080 |
| 15 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.433 |
0.075 |
| 16 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.429 |
0.165 |
| 17 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.413 |
0.166 |
| 18 |
260522_x_at |
AT2G41730
|
unknown |
0.409 |
0.010 |
| 19 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.407 |
0.086 |
| 20 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.378 |
-0.252 |
| 21 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.376 |
0.335 |
| 22 |
254403_at |
AT4G21323
|
[Subtilase family protein] |
0.375 |
0.089 |
| 23 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.369 |
0.038 |
| 24 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.364 |
-0.002 |
| 25 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.356 |
-0.183 |
| 26 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.354 |
0.249 |
| 27 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.348 |
-0.108 |
| 28 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.347 |
-0.053 |
| 29 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.336 |
0.132 |
| 30 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.334 |
0.201 |
| 31 |
248822_at |
AT5G47000
|
[Peroxidase superfamily protein] |
0.333 |
0.004 |
| 32 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.326 |
-0.281 |
| 33 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
0.320 |
0.103 |
| 34 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.319 |
0.144 |
| 35 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.319 |
0.120 |
| 36 |
249726_at |
AT5G35480
|
unknown |
0.312 |
-0.042 |
| 37 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.304 |
-0.231 |
| 38 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.303 |
-0.379 |
| 39 |
263851_at |
AT2G04460
|
unknown |
0.303 |
-0.114 |
| 40 |
250605_at |
AT5G07570
|
[glycine/proline-rich protein] |
0.303 |
0.092 |
| 41 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.299 |
0.094 |
| 42 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.297 |
0.331 |
| 43 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.293 |
0.130 |
| 44 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.291 |
-0.215 |
| 45 |
248752_at |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
0.284 |
0.006 |
| 46 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.284 |
0.214 |
| 47 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
0.280 |
0.221 |
| 48 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.277 |
0.225 |
| 49 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.275 |
-0.031 |
| 50 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.272 |
0.183 |
| 51 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.269 |
0.035 |
| 52 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.268 |
0.339 |
| 53 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.268 |
0.175 |
| 54 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.265 |
0.005 |
| 55 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.265 |
0.169 |
| 56 |
254263_at |
AT4G23493
|
unknown |
0.260 |
-0.013 |
| 57 |
258939_at |
AT3G10020
|
unknown |
0.258 |
0.047 |
| 58 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.248 |
0.126 |
| 59 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.232 |
0.200 |
| 60 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.232 |
0.026 |
| 61 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.231 |
-0.099 |
| 62 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.217 |
-0.122 |
| 63 |
245352_at |
AT4G15490
|
UGT84A3 |
0.216 |
-0.244 |
| 64 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.216 |
0.108 |
| 65 |
265441_at |
AT2G20870
|
[cell wall protein precursor, putative] |
0.215 |
0.118 |
| 66 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.215 |
0.032 |
| 67 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
0.214 |
0.119 |
| 68 |
258827_at |
AT3G07150
|
unknown |
0.210 |
0.051 |
| 69 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
0.208 |
-0.147 |
| 70 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.207 |
0.156 |
| 71 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.206 |
0.084 |
| 72 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.205 |
0.076 |
| 73 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
0.205 |
0.087 |
| 74 |
259554_at |
AT1G21220
|
[copia-like retrotransposon family, has a 3.9e-26 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
0.201 |
0.008 |
| 75 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
0.200 |
0.062 |
| 76 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
0.199 |
-0.128 |
| 77 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.199 |
-0.056 |
| 78 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
0.199 |
0.066 |
| 79 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.197 |
-0.034 |
| 80 |
250537_at |
AT5G08565
|
[Transcription initiation Spt4-like protein] |
0.196 |
0.090 |
| 81 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.195 |
0.106 |
| 82 |
257925_at |
AT3G23170
|
unknown |
0.193 |
0.141 |
| 83 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
0.192 |
-0.050 |
| 84 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
0.192 |
-0.052 |
| 85 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.192 |
0.299 |
| 86 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.192 |
0.132 |
| 87 |
257517_at |
AT3G16330
|
unknown |
0.192 |
0.186 |
| 88 |
259225_at |
AT3G07590
|
[Small nuclear ribonucleoprotein family protein] |
0.191 |
-0.007 |
| 89 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.190 |
0.081 |
| 90 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
0.189 |
0.138 |
| 91 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.189 |
-0.050 |
| 92 |
256363_at |
AT1G66510
|
[AAR2 protein family] |
0.188 |
0.011 |
| 93 |
249377_at |
AT5G40690
|
unknown |
0.187 |
0.002 |
| 94 |
257724_at |
AT3G18510
|
unknown |
0.186 |
-0.076 |
| 95 |
250639_at |
AT5G07560
|
ATGRP20, GRP20, glycine-rich protein 20 |
0.185 |
0.121 |
| 96 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.185 |
0.089 |
| 97 |
258406_at |
AT3G17611
|
ATRBL10, RHOMBOID-like protein 10, ATRBL14, RHOMBOID-like protein 14, RBL10, RHOMBOID-like protein 10, RBL14, RHOMBOID-like protein 14 |
0.185 |
0.129 |
| 98 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
0.182 |
0.093 |
| 99 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
0.181 |
0.012 |
| 100 |
258741_at |
AT3G05790
|
LON4, lon protease 4 |
0.181 |
0.010 |
| 101 |
259503_at |
AT1G15870
|
[Mitochondrial glycoprotein family protein] |
0.181 |
0.006 |
| 102 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.180 |
0.089 |
| 103 |
258452_at |
AT3G22370
|
AOX1A, alternative oxidase 1A, ATAOX1A, AtHSR3, HSR3, hyper-sensitivity-related 3 |
0.180 |
0.058 |
| 104 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
0.180 |
-0.103 |
| 105 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.180 |
0.084 |
| 106 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.179 |
-0.058 |
| 107 |
254200_at |
AT4G24110
|
unknown |
0.179 |
-0.061 |
| 108 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.178 |
0.046 |
| 109 |
255420_at |
AT4G03240
|
ATFH, FH, frataxin homolog |
0.178 |
0.046 |
| 110 |
247922_at |
AT5G57500
|
[Galactosyltransferase family protein] |
0.177 |
0.232 |
| 111 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.177 |
0.126 |
| 112 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
0.176 |
0.045 |
| 113 |
250224_at |
AT5G14150
|
unknown |
0.176 |
0.039 |
| 114 |
246612_at |
AT5G35320
|
unknown |
0.174 |
0.062 |
| 115 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.174 |
0.102 |
| 116 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.174 |
0.008 |
| 117 |
254786_at |
AT4G12890
|
[Gamma interferon responsive lysosomal thiol (GILT) reductase family protein] |
0.173 |
0.084 |
| 118 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.172 |
-0.012 |
| 119 |
250025_at |
AT5G18290
|
SIP1;2, SIP1B, SMALL AND BASIC INTRINSIC PROTEIN 1B |
0.172 |
0.145 |
| 120 |
245414_at |
AT4G17310
|
unknown |
0.171 |
0.120 |
| 121 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.171 |
0.071 |
| 122 |
259900_at |
AT1G71230
|
AJH2, CSN5, CSN5B, COP9-signalosome 5B |
0.171 |
0.106 |
| 123 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.170 |
-0.021 |
| 124 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.170 |
-0.708 |
| 125 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.170 |
0.433 |
| 126 |
259173_at |
AT3G03640
|
BGLU25, beta glucosidase 25, GLUC |
0.169 |
0.070 |
| 127 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
0.169 |
-0.079 |
| 128 |
245341_at |
AT4G16447
|
unknown |
0.168 |
0.003 |
| 129 |
246743_at |
AT5G27750
|
[F-box/FBD-like domains containing protein] |
0.168 |
0.048 |
| 130 |
260166_at |
AT1G79840
|
GL2, GLABRA 2 |
0.168 |
0.030 |
| 131 |
245903_at |
AT5G11100
|
ATSYTD, NTMC2T2.2, NTMC2TYPE2.2, SYT4, synaptotagmin 4, SYTD |
0.167 |
-0.015 |
| 132 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.167 |
0.303 |
| 133 |
248452_at |
AT5G51300
|
[splicing factor-related] |
-0.601 |
0.085 |
| 134 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-0.587 |
0.510 |
| 135 |
255418_at |
AT4G03200
|
[catalytics] |
-0.453 |
-0.191 |
| 136 |
262634_at |
AT1G06690
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.451 |
-0.164 |
| 137 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.415 |
0.029 |
| 138 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.415 |
-0.196 |
| 139 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.408 |
0.157 |
| 140 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.391 |
-0.181 |
| 141 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
-0.384 |
0.042 |
| 142 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.382 |
0.120 |
| 143 |
246411_at |
AT1G57770
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.360 |
-0.259 |
| 144 |
264270_at |
AT1G60260
|
BGLU5, beta glucosidase 5 |
-0.347 |
-0.182 |
| 145 |
267578_at |
AT2G30695
|
unknown |
-0.346 |
-0.343 |
| 146 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.345 |
0.121 |
| 147 |
256096_at |
AT1G13650
|
unknown |
-0.345 |
-0.647 |
| 148 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.341 |
-0.028 |
| 149 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.320 |
0.076 |
| 150 |
262648_at |
AT1G14030
|
LSMT-L, lysine methyltransferase (LSMT)-like |
-0.320 |
-0.280 |
| 151 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.309 |
0.214 |
| 152 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-0.301 |
0.074 |
| 153 |
251069_at |
AT5G01930
|
AtMAN6, MAN6, endo-beta-mannase 6 |
-0.279 |
-0.068 |
| 154 |
246951_at |
AT5G04880
|
[expressed protein] |
-0.277 |
-0.263 |
| 155 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.273 |
-0.011 |
| 156 |
261839_at |
AT1G16040
|
unknown |
-0.268 |
0.065 |
| 157 |
250809_at |
AT5G05140
|
[Transcription elongation factor (TFIIS) family protein] |
-0.268 |
0.194 |
| 158 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
-0.262 |
0.227 |
| 159 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.261 |
-0.270 |
| 160 |
265246_at |
AT2G43050
|
ATPMEPCRD |
-0.261 |
0.112 |
| 161 |
254817_at |
AT4G12460
|
ORP2B, OSBP(oxysterol binding protein)-related protein 2B |
-0.258 |
0.067 |
| 162 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
-0.258 |
0.068 |
| 163 |
257410_at |
AT1G24340
|
EMB2421, EMBRYO DEFECTIVE 2421, EMB260, EMBRYO DEFECTIVE 260 |
-0.257 |
-0.038 |
| 164 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
-0.255 |
0.239 |
| 165 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
-0.255 |
-0.479 |
| 166 |
257245_at |
AT3G24110
|
[Calcium-binding EF-hand family protein] |
-0.254 |
0.051 |
| 167 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.246 |
0.183 |
| 168 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.246 |
0.040 |
| 169 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
-0.245 |
0.139 |
| 170 |
250122_at |
AT5G16520
|
unknown |
-0.244 |
0.089 |
| 171 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
-0.242 |
0.129 |
| 172 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.238 |
-0.232 |
| 173 |
263410_at |
AT2G04039
|
unknown |
-0.237 |
-0.311 |
| 174 |
257447_at |
AT2G04230
|
[FBD, F-box and Leucine Rich Repeat domains containing protein] |
-0.232 |
0.033 |
| 175 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.232 |
0.181 |
| 176 |
245845_at |
AT1G26150
|
AtPERK10, proline-rich extensin-like receptor kinase 10, PERK10, proline-rich extensin-like receptor kinase 10 |
-0.232 |
-0.626 |
| 177 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.232 |
0.046 |
| 178 |
262561_at |
AT1G34340
|
[alpha/beta-Hydrolases superfamily protein] |
-0.231 |
0.189 |
| 179 |
251299_at |
AT3G61950
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.231 |
0.024 |
| 180 |
257761_at |
AT3G23090
|
WDL3 |
-0.231 |
0.094 |
| 181 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.229 |
0.033 |
| 182 |
257791_at |
AT3G27110
|
[Peptidase family M48 family protein] |
-0.226 |
-0.045 |
| 183 |
266689_at |
AT2G19930
|
[RNA-dependent RNA polymerase family protein] |
-0.226 |
-0.043 |
| 184 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.225 |
-0.053 |
| 185 |
264495_at |
AT1G27380
|
RIC2, ROP-interactive CRIB motif-containing protein 2 |
-0.223 |
0.064 |
| 186 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
-0.221 |
0.237 |
| 187 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
-0.221 |
-0.161 |
| 188 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.220 |
0.044 |
| 189 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
-0.219 |
0.024 |
| 190 |
249321_at |
AT5G40920
|
[pseudogene] |
-0.218 |
-0.042 |
| 191 |
253548_at |
AT4G30993
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.216 |
-0.547 |
| 192 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
-0.209 |
-0.281 |
| 193 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.208 |
-0.275 |
| 194 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.207 |
-0.032 |
| 195 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.207 |
0.044 |
| 196 |
257144_at |
AT3G27300
|
G6PD5, glucose-6-phosphate dehydrogenase 5 |
-0.207 |
0.132 |
| 197 |
267549_at |
AT2G32640
|
[Lycopene beta/epsilon cyclase protein] |
-0.205 |
-0.412 |
| 198 |
264622_at |
AT2G17790
|
VPS35A, VPS35 homolog A, ZIP3, ZIG suppressor 3 |
-0.205 |
0.161 |
| 199 |
259291_at |
AT3G11550
|
CASP2, Casparian strip membrane domain protein 2 |
-0.205 |
0.088 |
| 200 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.204 |
-0.078 |
| 201 |
245042_at |
AT2G26540
|
unknown |
-0.202 |
-0.074 |
| 202 |
251903_at |
AT3G54120
|
[Reticulon family protein] |
-0.198 |
0.092 |
| 203 |
249999_at |
AT5G18640
|
[alpha/beta-Hydrolases superfamily protein] |
-0.197 |
0.166 |
| 204 |
250861_at |
AT5G04740
|
ACR12, ACT domain repeats 12 |
-0.195 |
0.036 |
| 205 |
266625_at |
AT2G35380
|
[Peroxidase superfamily protein] |
-0.194 |
0.034 |
| 206 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.192 |
-0.241 |
| 207 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
-0.191 |
-0.225 |
| 208 |
254052_at |
AT4G25280
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.191 |
0.051 |
| 209 |
261066_at |
AT1G07485
|
unknown |
-0.190 |
0.016 |
| 210 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.190 |
-0.431 |
| 211 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.189 |
0.183 |
| 212 |
259862_at |
AT1G72650
|
TRFL6, TRF-like 6 |
-0.189 |
-0.119 |
| 213 |
260927_at |
AT1G05940
|
CAT9, cationic amino acid transporter 9 |
-0.188 |
0.024 |
| 214 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.188 |
-0.343 |
| 215 |
245730_at |
AT1G73470
|
unknown |
-0.187 |
-0.328 |
| 216 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.187 |
0.051 |
| 217 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
-0.187 |
-0.176 |
| 218 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.185 |
-0.164 |
| 219 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.183 |
0.009 |
| 220 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.182 |
0.060 |
| 221 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.182 |
-0.399 |
| 222 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.181 |
-0.067 |
| 223 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
-0.179 |
-0.242 |
| 224 |
262126_at |
AT1G59620
|
CW9 |
-0.179 |
0.054 |
| 225 |
257168_at |
AT3G24430
|
HCF101, HIGH-CHLOROPHYLL-FLUORESCENCE 101 |
-0.178 |
-0.515 |
| 226 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.178 |
0.148 |
| 227 |
255679_at |
AT4G00520
|
[Acyl-CoA thioesterase family protein] |
-0.177 |
0.026 |
| 228 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.177 |
-0.072 |
| 229 |
267081_at |
AT2G41210
|
PIP5K5, phosphatidylinositol- 4-phosphate 5-kinase 5 |
-0.175 |
0.021 |
| 230 |
248624_at |
AT5G48790
|
unknown |
-0.175 |
-0.127 |
| 231 |
262829_at |
AT1G14970
|
[O-fucosyltransferase family protein] |
-0.175 |
-0.012 |
| 232 |
258228_at |
AT3G27610
|
[Nucleotidylyl transferase superfamily protein] |
-0.175 |
0.003 |
| 233 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.175 |
-0.246 |
| 234 |
256619_at |
AT3G24460
|
[Serinc-domain containing serine and sphingolipid biosynthesis protein] |
-0.173 |
-0.064 |
| 235 |
246394_at |
AT1G58160
|
JAX1, JACALIN-TYPE LECTIN REQUIRED FOR POTEXVIRUS RESISTANCE 1 |
-0.173 |
0.100 |
| 236 |
266222_at |
AT2G28780
|
unknown |
-0.172 |
0.113 |
| 237 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.171 |
0.061 |
| 238 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.171 |
-0.129 |
| 239 |
262151_at |
AT1G52510
|
[alpha/beta-Hydrolases superfamily protein] |
-0.171 |
-0.254 |
| 240 |
250475_at |
AT5G10180
|
AST68, ARABIDOPSIS SULFATE TRANSPORTER 68, SULTR2;1, sulfate transporter 2;1 |
-0.170 |
0.030 |
| 241 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
-0.170 |
0.138 |
| 242 |
251119_at |
AT3G63510
|
[FMN-linked oxidoreductases superfamily protein] |
-0.170 |
-0.076 |
| 243 |
249162_at |
AT5G42765
|
unknown |
-0.170 |
-0.351 |
| 244 |
261382_at |
AT1G05470
|
CVP2, COTYLEDON VASCULAR PATTERN 2 |
-0.168 |
0.060 |
| 245 |
267569_at |
AT2G30790
|
PSBP-2, photosystem II subunit P-2 |
-0.167 |
0.071 |
| 246 |
248232_at |
AT5G53760
|
ATMLO11, MILDEW RESISTANCE LOCUS O 11, MLO11, MILDEW RESISTANCE LOCUS O 11 |
-0.167 |
-0.258 |
| 247 |
254622_at |
AT4G18375
|
[RNA-binding KH domain-containing protein] |
-0.166 |
0.072 |
| 248 |
252572_at |
AT3G45290
|
ATMLO3, MILDEW RESISTANCE LOCUS O 3, MLO3, MILDEW RESISTANCE LOCUS O 3 |
-0.166 |
0.083 |
| 249 |
254113_at |
AT4G24900
|
TTL, TITAN-like |
-0.165 |
0.066 |
| 250 |
256199_at |
AT1G58250
|
HPS4, hypersensitive to Pi starvation 4, SAB, SABRE |
-0.165 |
-0.042 |
| 251 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
-0.165 |
0.048 |
| 252 |
256541_at |
AT1G42540
|
ATGLR3.3, GLR3.3, glutamate receptor 3.3 |
-0.165 |
-0.011 |
| 253 |
266946_at |
AT2G18890
|
[Protein kinase superfamily protein] |
-0.164 |
0.118 |
| 254 |
251717_at |
AT3G55850
|
LAF3, LONG AFTER FAR-RED 3, LAF3 ISF1, LONG AFTER FAR-RED 3 ISOFORM 1, LAF3 ISF2, LAF3 ISOFORM 2 |
-0.164 |
0.149 |
| 255 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.163 |
0.006 |
| 256 |
256394_at |
AT3G06290
|
AtSAC3B, SAC3B, yeast Sac3 homolog B |
-0.162 |
0.031 |
| 257 |
251093_at |
AT5G01360
|
TBL3, TRICHOME BIREFRINGENCE-LIKE 3 |
-0.162 |
0.064 |
| 258 |
250728_at |
AT5G06440
|
unknown |
-0.161 |
-0.004 |
| 259 |
259258_at |
AT3G07670
|
[Rubisco methyltransferase family protein] |
-0.161 |
-0.420 |
| 260 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
-0.160 |
0.100 |
| 261 |
247223_at |
AT5G65070
|
AGL69, AGAMOUS-like 69, FCL4, MAF4, MADS AFFECTING FLOWERING 4 |
-0.160 |
0.023 |
| 262 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.160 |
-0.137 |
| 263 |
249006_at |
AT5G44660
|
unknown |
-0.160 |
-0.076 |