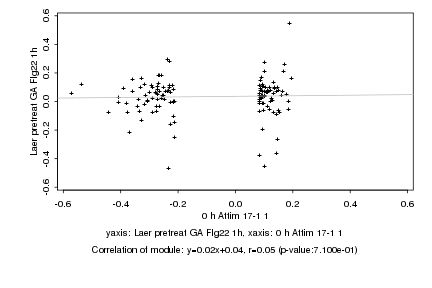
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
0 h Attim 17-1 1 |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
260909_at |
AT1G02670
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.195 |
0.162 |
| 2 |
261466_at |
AT1G07690
|
unknown |
0.186 |
0.552 |
| 3 |
248488_at |
AT5G51080
|
[RNase H family protein] |
0.184 |
-0.049 |
| 4 |
263230_at |
AT1G05670
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.183 |
0.007 |
| 5 |
265296_at |
AT2G14060
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.176 |
0.054 |
| 6 |
260256_at |
AT1G74350
|
[Intron maturase, type II family protein] |
0.169 |
0.264 |
| 7 |
251130_at |
AT5G01180
|
AtNPF8.2, ATPTR5, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 5, NPF8.2, NRT1/ PTR family 8.2, PTR5, peptide transporter 5 |
0.165 |
0.212 |
| 8 |
258248_at |
AT3G29140
|
unknown |
0.161 |
0.074 |
| 9 |
253671_at |
AT4G29990
|
[Leucine-rich repeat transmembrane protein kinase protein] |
0.160 |
0.044 |
| 10 |
258324_at |
AT3G22780
|
ATTSO1, TSO1, CHINESE FOR 'UGLY' |
0.152 |
-0.074 |
| 11 |
248753_at |
AT5G47630
|
mtACP3, mitochondrial acyl carrier protein 3 |
0.149 |
-0.060 |
| 12 |
249897_at |
AT5G22550
|
unknown |
0.147 |
0.079 |
| 13 |
261801_at |
AT1G30520
|
AAE14, acyl-activating enzyme 14 |
0.145 |
-0.262 |
| 14 |
249816_at |
AT5G23880
|
ATCPSF100, CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 100, CPSF100, cleavage and polyadenylation specificity factor 100, EMB1265, EMBRYO DEFECTIVE 1265, ESP5, ENHANCED SILENCING PHENOTYPE 5 |
0.145 |
0.102 |
| 15 |
259797_at |
AT1G64410
|
unknown |
0.142 |
0.073 |
| 16 |
252282_at |
AT3G49360
|
PGL2, 6-phosphogluconolactonase 2 |
0.141 |
-0.361 |
| 17 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
0.140 |
-0.084 |
| 18 |
247911_at |
AT5G57450
|
ATXRCC3, ARABIDOPSIS THALIANA HOMOLOG OF X-RAY REPAIR CROSS COMPLEMENTING 3 (XRCC3), XRCC3, homolog of X-ray repair cross complementing 3 |
0.135 |
0.102 |
| 19 |
244905_at |
ATMG00680
|
ORF122C |
0.134 |
0.097 |
| 20 |
253413_at |
AT4G33020
|
ATZIP9, ZIP9 |
0.132 |
0.137 |
| 21 |
252530_at |
AT3G46500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.131 |
-0.074 |
| 22 |
245520_at |
AT4G15870
|
ATTS1, terpene synthase 1, TS1, terpene synthase 1 |
0.131 |
0.057 |
| 23 |
257133_at |
AT3G17190
|
unknown |
0.127 |
0.010 |
| 24 |
256640_at |
AT3G32260
|
[Nucleic acid-binding proteins superfamily] |
0.125 |
0.025 |
| 25 |
256685_at |
AT3G32100
|
unknown |
0.121 |
0.078 |
| 26 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
0.121 |
0.005 |
| 27 |
247578_at |
AT5G61300
|
unknown |
0.118 |
-0.053 |
| 28 |
251530_at |
AT3G58520
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.116 |
0.102 |
| 29 |
253074_at |
AT4G36140
|
[disease resistance protein (TIR-NBS-LRR class), putative] |
0.112 |
0.076 |
| 30 |
267091_at |
AT2G38185
|
APD1, ABERRANT POLLEN DEVELOPMENT 1 |
0.110 |
-0.029 |
| 31 |
263974_at |
AT2G42720
|
[FBD, F-box, Skp2-like and Leucine Rich Repeat domains containing protein] |
0.110 |
0.070 |
| 32 |
257531_at |
AT3G07250
|
[nuclear transport factor 2 (NTF2) family protein / RNA recognition motif (RRM)-containing protein] |
0.109 |
0.080 |
| 33 |
252732_at |
AT3G43140
|
transposable element gene |
0.108 |
0.067 |
| 34 |
260437_at |
AT1G68380
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.104 |
0.103 |
| 35 |
254589_at |
AT4G18850
|
unknown |
0.101 |
0.043 |
| 36 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
0.101 |
0.217 |
| 37 |
257773_at |
AT3G29185
|
unknown |
0.101 |
-0.452 |
| 38 |
262276_at |
AT1G68700
|
unknown |
0.100 |
0.074 |
| 39 |
248454_at |
AT5G51350
|
MOL1, MORE LATERAL GROWTH1 |
0.099 |
0.041 |
| 40 |
247547_at |
AT5G61360
|
unknown |
0.098 |
0.277 |
| 41 |
261915_at |
AT1G65880
|
BZO1, benzoyloxyglucosinolate 1 |
0.097 |
0.112 |
| 42 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.096 |
-0.056 |
| 43 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.095 |
-0.012 |
| 44 |
263052_at |
AT2G13430
|
unknown |
0.094 |
0.047 |
| 45 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.093 |
-0.191 |
| 46 |
256428_at |
AT3G11080
|
AtRLP35, receptor like protein 35, RLP35, receptor like protein 35 |
0.093 |
0.121 |
| 47 |
246662_at |
AT5G35290
|
unknown |
0.093 |
0.074 |
| 48 |
245680_at |
AT1G56570
|
PGN, PENTATRICOPEPTIDE REPEAT PROTEIN FOR GERMINATION ON NaCl |
0.093 |
-0.013 |
| 49 |
254401_at |
AT4G21300
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.093 |
0.109 |
| 50 |
258255_at |
AT3G26800
|
unknown |
0.090 |
0.171 |
| 51 |
265694_at |
AT2G24440
|
[selenium binding] |
0.090 |
0.082 |
| 52 |
265140_at |
AT1G51320
|
[F-box and associated interaction domains-containing protein] |
0.089 |
0.023 |
| 53 |
267331_at |
AT2G19280
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.088 |
0.044 |
| 54 |
263266_at |
AT2G16520
|
unknown |
0.088 |
0.031 |
| 55 |
247017_at |
AT5G66980
|
[AP2/B3-like transcriptional factor family protein] |
0.087 |
0.097 |
| 56 |
250251_at |
AT5G13670
|
UMAMIT15, Usually multiple acids move in and out Transporters 15 |
0.085 |
0.078 |
| 57 |
252659_at |
AT3G44430
|
unknown |
0.085 |
0.151 |
| 58 |
247367_at |
AT5G63290
|
[Radical SAM superfamily protein] |
0.085 |
0.029 |
| 59 |
250745_at |
AT5G05850
|
PIRL1, plant intracellular ras group-related LRR 1 |
0.083 |
0.113 |
| 60 |
252541_at |
AT3G45750
|
[Nucleotidyltransferase family protein] |
0.083 |
0.021 |
| 61 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
0.082 |
-0.011 |
| 62 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.082 |
-0.371 |
| 63 |
265371_at |
AT2G06490
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.5e-83 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.082 |
0.047 |
| 64 |
260422_at |
AT1G69630
|
[F-box/RNI-like superfamily protein] |
0.081 |
0.059 |
| 65 |
266333_at |
AT2G32410
|
AXL, AXR1-like, AXL1, AXR1- like 1 |
0.081 |
0.006 |
| 66 |
247596_at |
AT5G60840
|
unknown |
0.081 |
-0.066 |
| 67 |
265127_at |
AT1G55560
|
sks14, SKU5 similar 14 |
-0.574 |
0.062 |
| 68 |
251635_at |
AT3G57510
|
ADPG1 |
-0.538 |
0.122 |
| 69 |
264545_at |
AT1G55670
|
PSAG, photosystem I subunit G |
-0.444 |
-0.073 |
| 70 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.411 |
0.030 |
| 71 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.409 |
-0.005 |
| 72 |
257832_at |
AT3G26740
|
CCL, CCR-like |
-0.393 |
0.098 |
| 73 |
248470_at |
AT5G50830
|
unknown |
-0.382 |
-0.010 |
| 74 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.377 |
-0.073 |
| 75 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
-0.370 |
-0.210 |
| 76 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.359 |
0.157 |
| 77 |
266115_at |
AT2G02140
|
LCR72, low-molecular-weight cysteine-rich 72, PDF2.6 |
-0.359 |
0.072 |
| 78 |
260969_at |
AT1G12240
|
ATBETAFRUCT4, AtFRUCT4, AtVI2, FRUCT4, fructosidase 4, VAC-INV, VACUOLAR INVERTASE, VI2, vacuolar invertase 2 |
-0.342 |
-0.034 |
| 79 |
258889_at |
AT3G05610
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.341 |
0.016 |
| 80 |
265033_at |
AT1G61520
|
LHCA3, photosystem I light harvesting complex gene 3 |
-0.336 |
-0.068 |
| 81 |
247512_at |
AT5G61720
|
unknown |
-0.331 |
0.102 |
| 82 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
-0.329 |
0.166 |
| 83 |
259570_at |
AT1G20440
|
AtCOR47, COR47, cold-regulated 47, RD17 |
-0.328 |
-0.132 |
| 84 |
262958_at |
AT1G54410
|
AtHIRD11, HIRD11, dehydrin 11kDa |
-0.318 |
-0.015 |
| 85 |
246841_at |
AT5G26700
|
[RmlC-like cupins superfamily protein] |
-0.318 |
0.121 |
| 86 |
253226_at |
AT4G35010
|
BGAL11, beta-galactosidase 11 |
-0.314 |
0.043 |
| 87 |
248822_at |
AT5G47000
|
[Peroxidase superfamily protein] |
-0.310 |
0.004 |
| 88 |
256581_at |
AT3G28830
|
unknown |
-0.307 |
0.011 |
| 89 |
252348_at |
AT3G48140
|
[B12D protein] |
-0.301 |
0.066 |
| 90 |
253277_at |
AT4G34230
|
ATCAD5, cinnamyl alcohol dehydrogenase 5, CAD-5, cinnamyl alcohol dehydrogenase 5, CAD5, cinnamyl alcohol dehydrogenase 5 |
-0.295 |
0.119 |
| 91 |
251733_at |
AT3G56240
|
CCH, copper chaperone |
-0.292 |
-0.073 |
| 92 |
265080_at |
AT1G55570
|
sks12, SKU5 similar 12 |
-0.292 |
0.026 |
| 93 |
251228_at |
AT3G62710
|
[Glycosyl hydrolase family protein] |
-0.290 |
0.104 |
| 94 |
245658_at |
AT1G28270
|
AtRALF4, RALFL4, ralf-like 4 |
-0.281 |
0.065 |
| 95 |
263688_at |
AT1G26920
|
unknown |
-0.278 |
-0.033 |
| 96 |
250154_at |
AT5G15140
|
[Galactose mutarotase-like superfamily protein] |
-0.277 |
0.063 |
| 97 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
-0.277 |
-0.067 |
| 98 |
249852_at |
AT5G23270
|
ATSTP11, SUGAR TRANSPORTER 11, STP11, sugar transporter 11 |
-0.275 |
0.087 |
| 99 |
252024_at |
AT3G52880
|
ATMDAR1, monodehydroascorbate reductase 1, MDAR1, monodehydroascorbate reductase 1 |
-0.274 |
0.050 |
| 100 |
260004_at |
AT1G67860
|
unknown |
-0.273 |
0.107 |
| 101 |
245803_at |
AT1G47128
|
RD21, responsive to dehydration 21, RD21A, responsive to dehydration 21A |
-0.272 |
0.017 |
| 102 |
266918_at |
AT2G45800
|
PLIM2a, PLIM2a |
-0.269 |
0.129 |
| 103 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
-0.269 |
0.185 |
| 104 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
-0.268 |
-0.030 |
| 105 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
-0.266 |
0.184 |
| 106 |
261412_at |
AT1G07890
|
APX1, ascorbate peroxidase 1, ATAPX01, ATAPX1, CS1, MEE6, maternal effect embryo arrest 6 |
-0.266 |
0.073 |
| 107 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.260 |
0.027 |
| 108 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
-0.258 |
0.183 |
| 109 |
261528_at |
AT1G14420
|
AT59 |
-0.255 |
0.043 |
| 110 |
256588_at |
AT3G28790
|
unknown |
-0.254 |
0.101 |
| 111 |
261623_at |
AT1G01980
|
[ATSEC1A] |
-0.253 |
0.049 |
| 112 |
258639_at |
AT3G07820
|
[Pectin lyase-like superfamily protein] |
-0.250 |
0.016 |
| 113 |
266764_at |
AT2G47050
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.247 |
0.077 |
| 114 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
-0.240 |
0.295 |
| 115 |
256966_at |
AT3G13400
|
sks13, SKU5 similar 13 |
-0.238 |
0.074 |
| 116 |
261749_at |
AT1G76180
|
ERD14, EARLY RESPONSE TO DEHYDRATION 14 |
-0.236 |
0.080 |
| 117 |
262609_at |
AT1G13930
|
unknown |
-0.234 |
-0.467 |
| 118 |
250631_at |
AT5G07430
|
[Pectin lyase-like superfamily protein] |
-0.233 |
0.114 |
| 119 |
259189_at |
AT3G01700
|
AGP11, arabinogalactan protein 11, ATAGP11, ARABINOGALACTAN PROTEIN 11 |
-0.233 |
0.103 |
| 120 |
251281_at |
AT3G61640
|
AGP20, arabinogalactan protein 20, AtAGP20, Arabinogalactan protein 20 |
-0.232 |
0.281 |
| 121 |
259269_at |
AT3G01270
|
[Pectate lyase family protein] |
-0.229 |
-0.003 |
| 122 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
-0.229 |
-0.157 |
| 123 |
257771_at |
AT3G23000
|
ATSR2, ATSRPK1, CIPK7, CBL-interacting protein kinase 7, PKS7, SnRK3.10, SNF1-RELATED PROTEIN KINASE 3.10 |
-0.227 |
0.070 |
| 124 |
245700_at |
AT5G04180
|
ACA3, alpha carbonic anhydrase 3, ATACA3, ALPHA CARBONIC ANHYDRASE 3 |
-0.221 |
0.116 |
| 125 |
245946_at |
AT5G19580
|
[glyoxal oxidase-related protein] |
-0.218 |
0.005 |
| 126 |
257986_at |
AT3G20865
|
AGP40, arabinogalactan protein 40 |
-0.218 |
0.087 |
| 127 |
253738_at |
AT4G28750
|
PSAE-1, PSA E1 KNOCKOUT |
-0.218 |
-0.100 |
| 128 |
259886_at |
AT1G76370
|
[Protein kinase superfamily protein] |
-0.217 |
-0.002 |
| 129 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.215 |
-0.142 |
| 130 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
-0.215 |
-0.250 |
| 131 |
262786_at |
AT1G10740
|
[alpha/beta-Hydrolases superfamily protein] |
-0.215 |
0.001 |