|
probeID |
AGICode |
Annotation |
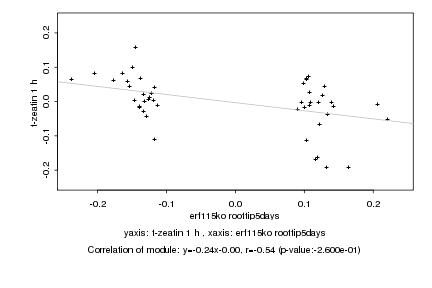
Log2 signal ratio
erf115ko roottip5days |
Log2 signal ratio
t-zeatin 1 h |
| 1 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.220 |
-0.052 |
| 2 |
245532_at |
AT4G15110
|
CYP97B3, cytochrome P450, family 97, subfamily B, polypeptide 3 |
0.205 |
-0.006 |
| 3 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.163 |
-0.192 |
| 4 |
249927_at |
AT5G19220
|
ADG2, ADP GLUCOSE PYROPHOSPHORYLASE 2, APL1, ADP glucose pyrophosphorylase large subunit 1 |
0.141 |
-0.013 |
| 5 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.138 |
-0.002 |
| 6 |
262449_at |
AT1G11160
|
[Transducin/WD40 repeat-like superfamily protein] |
0.133 |
-0.036 |
| 7 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.132 |
-0.192 |
| 8 |
251871_at |
AT3G54520
|
unknown |
0.128 |
0.045 |
| 9 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.125 |
0.020 |
| 10 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.122 |
-0.065 |
| 11 |
264278_at |
AT1G60130
|
[Mannose-binding lectin superfamily protein] |
0.120 |
-0.001 |
| 12 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.118 |
-0.163 |
| 13 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.115 |
-0.167 |
| 14 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
0.108 |
-0.001 |
| 15 |
256247_at |
AT3G66656
|
AGL91, AGAMOUS-like 91 |
0.107 |
-0.011 |
| 16 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.107 |
0.029 |
| 17 |
248832_at |
AT5G47170
|
unknown |
0.105 |
0.073 |
| 18 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.103 |
-0.113 |
| 19 |
255903_at |
AT1G17950
|
ATMYB52, MYB DOMAIN PROTEIN 52, BW52, MYB52, myb domain protein 52 |
0.103 |
0.065 |
| 20 |
246077_at |
AT5G20420
|
CHR42, chromatin remodeling 42 |
0.102 |
0.068 |
| 21 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
0.099 |
-0.016 |
| 22 |
264948_at |
AT1G77030
|
[hydrolases, acting on acid anhydrides, in phosphorus-containing anhydrides] |
0.098 |
0.055 |
| 23 |
244934_at |
ATCG01080
|
NDHG |
0.095 |
-0.000 |
| 24 |
256992_at |
AT3G28630
|
CROLIN1, CROLIN 1 |
0.090 |
-0.023 |
| 25 |
250616_at |
AT5G07280
|
EMS1, EXCESS MICROSPOROCYTES1, EXS, EXTRA SPOROGENOUS CELLS |
-0.239 |
0.066 |
| 26 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
-0.205 |
0.083 |
| 27 |
251377_at |
AT3G60650
|
unknown |
-0.177 |
0.064 |
| 28 |
256009_at |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
-0.165 |
0.083 |
| 29 |
262833_at |
AT1G14750
|
SDS, SOLO DANCERS |
-0.158 |
0.060 |
| 30 |
257804_at |
AT3G18810
|
AtPERK6, proline-rich extensin-like receptor kinase 6, PERK6, proline-rich extensin-like receptor kinase 6 |
-0.154 |
0.045 |
| 31 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.150 |
0.100 |
| 32 |
246107_at |
AT5G28700
|
[pseudogene] |
-0.147 |
0.004 |
| 33 |
259850_at |
AT1G72240
|
unknown |
-0.146 |
0.158 |
| 34 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
-0.140 |
-0.017 |
| 35 |
255316_at |
AT4G04170
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.3e-87 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.140 |
-0.014 |
| 36 |
259820_at |
AT1G66210
|
[Subtilisin-like serine endopeptidase family protein] |
-0.138 |
0.070 |
| 37 |
262350_at |
AT2G48150
|
ATGPX4, glutathione peroxidase 4, GPX4, glutathione peroxidase 4 |
-0.134 |
0.023 |
| 38 |
254719_at |
AT4G13600
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.134 |
-0.028 |
| 39 |
255023_at |
AT4G09850
|
unknown |
-0.132 |
0.002 |
| 40 |
248868_at |
AT5G46780
|
[VQ motif-containing protein] |
-0.130 |
-0.043 |
| 41 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-0.127 |
0.006 |
| 42 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
-0.126 |
0.013 |
| 43 |
264909_at |
AT2G17300
|
unknown |
-0.123 |
0.026 |
| 44 |
253284_at |
AT4G34150
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.120 |
0.005 |
| 45 |
253179_at |
AT4G35200
|
unknown |
-0.118 |
0.043 |
| 46 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.118 |
-0.109 |
| 47 |
264632_at |
AT1G65640
|
DEG4, degradation of periplasmic proteins 4, DegP4, DegP protease 4 |
-0.114 |
-0.011 |