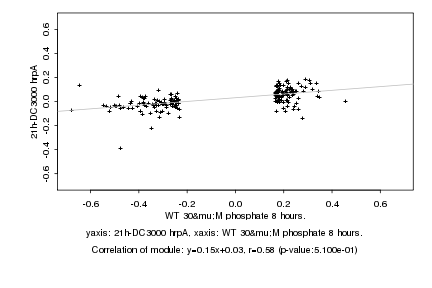
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT 30μM phosphate 8 hours. |
Log2 signal ratio
21h-DC3000 hrpA |
| 1 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
0.452 |
0.001 |
| 2 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.348 |
0.040 |
| 3 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.341 |
0.086 |
| 4 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
0.338 |
0.042 |
| 5 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
0.334 |
0.154 |
| 6 |
246303_at |
AT3G51870
|
[Mitochondrial substrate carrier family protein] |
0.316 |
0.103 |
| 7 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.309 |
0.156 |
| 8 |
256675_at |
AT3G52170
|
[DNA binding] |
0.303 |
0.180 |
| 9 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.289 |
0.186 |
| 10 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
0.287 |
0.118 |
| 11 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
0.279 |
0.090 |
| 12 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
0.274 |
-0.140 |
| 13 |
262706_at |
AT1G16280
|
AtRH36, Arabidopsis thaliana RNA helicase 36, RH36, RNA helicase 36, SWA3, SLOW WALKER 3 |
0.270 |
0.125 |
| 14 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.261 |
0.032 |
| 15 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
0.261 |
0.152 |
| 16 |
250936_at |
AT5G03120
|
unknown |
0.258 |
-0.063 |
| 17 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
0.252 |
-0.016 |
| 18 |
262766_at |
AT1G13160
|
[ARM repeat superfamily protein] |
0.252 |
0.088 |
| 19 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.247 |
0.086 |
| 20 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.244 |
-0.041 |
| 21 |
258316_at |
AT3G22660
|
[rRNA processing protein-related] |
0.240 |
0.085 |
| 22 |
258202_at |
AT3G13940
|
[DNA binding] |
0.239 |
0.065 |
| 23 |
252508_at |
AT3G46210
|
[Ribosomal protein S5 domain 2-like superfamily protein] |
0.238 |
0.079 |
| 24 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
0.237 |
-0.064 |
| 25 |
266801_at |
AT2G22870
|
EMB2001, embryo defective 2001 |
0.236 |
0.057 |
| 26 |
247942_at |
AT5G57120
|
unknown |
0.235 |
0.077 |
| 27 |
267309_at |
AT2G19385
|
[zinc ion binding] |
0.234 |
0.107 |
| 28 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
0.231 |
0.089 |
| 29 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.226 |
0.077 |
| 30 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.225 |
0.109 |
| 31 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.225 |
0.117 |
| 32 |
259672_at |
AT1G68990
|
MGP3, male gametophyte defective 3 |
0.220 |
0.103 |
| 33 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
0.220 |
0.041 |
| 34 |
265326_at |
AT2G18220
|
[Noc2p family] |
0.218 |
0.153 |
| 35 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
0.216 |
-0.007 |
| 36 |
246457_at |
AT5G16750
|
TOZ, TORMOZEMBRYO DEFECTIVE |
0.215 |
0.106 |
| 37 |
247032_at |
AT5G67240
|
SDN3, small RNA degrading nuclease 3 |
0.215 |
0.123 |
| 38 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.214 |
0.053 |
| 39 |
245909_at |
AT5G09380
|
[RNA polymerase III RPC4] |
0.214 |
0.012 |
| 40 |
247926_at |
AT5G57280
|
RID2, root initiation defective 2 |
0.213 |
0.064 |
| 41 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.212 |
0.180 |
| 42 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
0.212 |
-0.039 |
| 43 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
0.211 |
0.174 |
| 44 |
263255_at |
AT1G10490
|
unknown |
0.210 |
0.016 |
| 45 |
261301_at |
AT1G48570
|
[zinc finger (Ran-binding) family protein] |
0.209 |
0.096 |
| 46 |
267371_at |
AT2G44510
|
[CDK inhibitor P21 binding protein] |
0.208 |
0.079 |
| 47 |
257571_at |
AT3G16870
|
GATA17, GATA transcription factor 17 |
0.208 |
0.064 |
| 48 |
247754_at |
AT5G59080
|
unknown |
0.207 |
-0.082 |
| 49 |
260316_at |
AT1G63810
|
unknown |
0.205 |
0.090 |
| 50 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.199 |
-0.005 |
| 51 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.198 |
-0.055 |
| 52 |
248357_at |
AT5G52380
|
[VASCULAR-RELATED NAC-DOMAIN 6] |
0.197 |
0.056 |
| 53 |
266076_at |
AT2G40700
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.197 |
0.073 |
| 54 |
253005_at |
AT4G38100
|
CURT1D, CURVATURE THYLAKOID 1D |
0.197 |
0.070 |
| 55 |
245518_at |
AT4G15850
|
ATRH1, RNA helicase 1, RH1, RNA helicase 1 |
0.195 |
0.140 |
| 56 |
258794_at |
AT3G04710
|
TPR10, tetratricopeptide repeat 10 |
0.194 |
0.086 |
| 57 |
247497_at |
AT5G61770
|
PPAN, PETER PAN-like protein |
0.193 |
0.079 |
| 58 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
0.192 |
0.056 |
| 59 |
248036_at |
AT5G55920
|
OLI2, OLIGOCELLULA 2 |
0.191 |
0.058 |
| 60 |
258630_at |
AT3G02820
|
[zinc knuckle (CCHC-type) family protein] |
0.188 |
0.015 |
| 61 |
262207_at |
AT1G74850
|
PDE343, PIGMENT DEFECTIVE 343, PTAC2, plastid transcriptionally active 2 |
0.187 |
0.041 |
| 62 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
0.183 |
0.138 |
| 63 |
247235_at |
AT5G64580
|
EMB3144, EMBRYO DEFECTIVE 3144 |
0.182 |
0.094 |
| 64 |
256284_at |
AT3G12530
|
PSF2 |
0.181 |
0.151 |
| 65 |
262738_at |
AT1G28530
|
unknown |
0.181 |
0.046 |
| 66 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.181 |
0.024 |
| 67 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.179 |
0.010 |
| 68 |
263864_at |
AT2G04530
|
CPZ, TRZ2, TRNASE Z 2 |
0.179 |
0.115 |
| 69 |
267445_at |
AT2G33680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.179 |
-0.007 |
| 70 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.178 |
0.094 |
| 71 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
0.178 |
0.003 |
| 72 |
259367_at |
AT1G69070
|
unknown |
0.178 |
0.036 |
| 73 |
256107_at |
AT1G16830
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.178 |
0.104 |
| 74 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
0.178 |
0.080 |
| 75 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
0.175 |
0.174 |
| 76 |
254092_at |
AT4G25090
|
[Riboflavin synthase-like superfamily protein] |
0.174 |
-0.002 |
| 77 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.173 |
0.126 |
| 78 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
0.172 |
0.138 |
| 79 |
264002_at |
AT2G22360
|
DJA6, DNA J protein A6 |
0.172 |
0.003 |
| 80 |
267044_at |
AT2G34357
|
[ARM repeat superfamily protein] |
0.171 |
0.076 |
| 81 |
262272_at |
AT1G42440
|
unknown |
0.171 |
0.051 |
| 82 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
0.170 |
0.133 |
| 83 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
0.170 |
0.133 |
| 84 |
260425_at |
AT1G72440
|
EDA25, embryo sac development arrest 25, SWA2, SLOW WALKER2 |
0.170 |
0.141 |
| 85 |
246350_at |
AT1G16650
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.169 |
0.063 |
| 86 |
255749_at |
AT1G31970
|
STRS1, STRESS RESPONSE SUPPRESSOR 1 |
0.169 |
0.042 |
| 87 |
247046_at |
AT5G66540
|
unknown |
0.168 |
0.006 |
| 88 |
256661_at |
AT3G11964
|
RRP5, Ribosomal RNA Processing 5 |
0.166 |
0.087 |
| 89 |
248443_at |
AT5G51310
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.166 |
-0.075 |
| 90 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
0.165 |
0.026 |
| 91 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.164 |
0.008 |
| 92 |
249433_at |
AT5G39940
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.164 |
0.069 |
| 93 |
261168_at |
AT1G04945
|
[HIT-type Zinc finger family protein] |
0.163 |
0.052 |
| 94 |
259908_at |
AT1G60850
|
AAC42, ATRPAC42 |
0.163 |
0.079 |
| 95 |
258872_at |
AT3G03260
|
HDG8, homeodomain GLABROUS 8 |
-0.682 |
-0.069 |
| 96 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.646 |
0.137 |
| 97 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
-0.549 |
-0.033 |
| 98 |
254111_at |
AT4G24890
|
ATPAP24, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 24, PAP24, purple acid phosphatase 24 |
-0.535 |
-0.035 |
| 99 |
259974_at |
AT1G76430
|
PHT1;9, phosphate transporter 1;9 |
-0.525 |
-0.077 |
| 100 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.521 |
-0.042 |
| 101 |
250091_at |
AT5G17340
|
[Putative membrane lipoprotein] |
-0.502 |
-0.029 |
| 102 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.499 |
-0.040 |
| 103 |
254097_at |
AT4G25160
|
[U-box domain-containing protein kinase family protein] |
-0.495 |
-0.034 |
| 104 |
253087_at |
AT4G36350
|
ATPAP25, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 25, PAP25, purple acid phosphatase 25 |
-0.486 |
0.047 |
| 105 |
258172_at |
AT3G21620
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.483 |
-0.025 |
| 106 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
-0.479 |
-0.056 |
| 107 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
-0.478 |
-0.389 |
| 108 |
252730_at |
AT3G43110
|
unknown |
-0.465 |
-0.048 |
| 109 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
-0.447 |
-0.052 |
| 110 |
260148_at |
AT1G52800
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.435 |
-0.015 |
| 111 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.435 |
-0.009 |
| 112 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.433 |
0.005 |
| 113 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.428 |
-0.055 |
| 114 |
262658_at |
AT1G14220
|
[Ribonuclease T2 family protein] |
-0.409 |
-0.041 |
| 115 |
254729_at |
AT4G13700
|
ATPAP23, PAP23, purple acid phosphatase 23 |
-0.400 |
-0.015 |
| 116 |
254096_at |
AT4G25150
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.397 |
0.043 |
| 117 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.396 |
-0.080 |
| 118 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.394 |
-0.076 |
| 119 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.387 |
0.038 |
| 120 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.385 |
-0.104 |
| 121 |
251315_at |
AT3G61410
|
unknown |
-0.383 |
-0.013 |
| 122 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
-0.381 |
0.028 |
| 123 |
265387_at |
AT2G20670
|
unknown |
-0.381 |
-0.002 |
| 124 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-0.380 |
-0.033 |
| 125 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
-0.380 |
0.033 |
| 126 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
-0.376 |
0.049 |
| 127 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
-0.371 |
-0.038 |
| 128 |
260522_x_at |
AT2G41730
|
unknown |
-0.361 |
-0.010 |
| 129 |
254290_at |
AT4G23000
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.353 |
-0.092 |
| 130 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.348 |
-0.222 |
| 131 |
253474_at |
AT4G32270
|
[Ubiquitin-like superfamily protein] |
-0.346 |
-0.018 |
| 132 |
249747_at |
AT5G24600
|
unknown |
-0.341 |
-0.028 |
| 133 |
246551_at |
AT5G15070
|
[Phosphoglycerate mutase-like family protein] |
-0.337 |
0.024 |
| 134 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.336 |
-0.044 |
| 135 |
247287_at |
AT5G64230
|
unknown |
-0.334 |
-0.011 |
| 136 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
-0.330 |
-0.076 |
| 137 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
-0.330 |
-0.015 |
| 138 |
259750_at |
AT1G71130
|
AtERF070, CRF8, cytokinin response factor 8, ERF070, ethylene response factor 70 |
-0.328 |
-0.031 |
| 139 |
261817_at |
AT1G08180
|
unknown |
-0.324 |
0.015 |
| 140 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
-0.321 |
0.094 |
| 141 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
-0.319 |
-0.027 |
| 142 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.317 |
-0.127 |
| 143 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
-0.317 |
0.005 |
| 144 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
-0.312 |
-0.084 |
| 145 |
253296_at |
AT4G33770
|
ITPK2, inositol 1,3,4-trisphosphate 5/6 kinase 2 |
-0.310 |
-0.008 |
| 146 |
255900_at |
AT1G17830
|
unknown |
-0.306 |
-0.077 |
| 147 |
246045_at |
AT5G19430
|
[RING/U-box superfamily protein] |
-0.306 |
-0.019 |
| 148 |
256014_at |
AT1G19200
|
unknown |
-0.305 |
-0.017 |
| 149 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.303 |
-0.019 |
| 150 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
-0.301 |
-0.021 |
| 151 |
249721_at |
AT5G24655
|
LSU4, RESPONSE TO LOW SULFUR 4 |
-0.295 |
-0.003 |
| 152 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.294 |
0.018 |
| 153 |
251741_at |
AT3G56040
|
UGP3, UDP-glucose pyrophosphorylase 3 |
-0.292 |
-0.018 |
| 154 |
245882_at |
AT5G09470
|
DIC3, dicarboxylate carrier 3 |
-0.286 |
-0.032 |
| 155 |
255763_at |
AT1G16730
|
unknown |
-0.286 |
-0.042 |
| 156 |
266564_at |
AT2G24000
|
scpl22, serine carboxypeptidase-like 22 |
-0.281 |
-0.098 |
| 157 |
262411_at |
AT1G34640
|
[peptidases] |
-0.270 |
0.014 |
| 158 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
-0.269 |
0.065 |
| 159 |
259334_at |
AT3G03790
|
[ankyrin repeat family protein / regulator of chromosome condensation (RCC1) family protein] |
-0.269 |
-0.021 |
| 160 |
245495_at |
AT4G16400
|
unknown |
-0.268 |
0.030 |
| 161 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
-0.268 |
0.063 |
| 162 |
267423_at |
AT2G35060
|
KUP11, K+ uptake permease 11 |
-0.267 |
-0.005 |
| 163 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.266 |
0.020 |
| 164 |
254796_at |
AT4G13000
|
[AGC (cAMP-dependent, cGMP-dependent and protein kinase C) kinase family protein] |
-0.262 |
-0.007 |
| 165 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
-0.261 |
-0.024 |
| 166 |
248218_at |
AT5G53710
|
unknown |
-0.261 |
-0.041 |
| 167 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
-0.260 |
-0.005 |
| 168 |
245082_at |
AT2G23270
|
unknown |
-0.257 |
0.006 |
| 169 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
-0.254 |
0.016 |
| 170 |
266100_at |
AT2G37980
|
[O-fucosyltransferase family protein] |
-0.254 |
-0.040 |
| 171 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-0.251 |
-0.002 |
| 172 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.251 |
-0.045 |
| 173 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.251 |
-0.027 |
| 174 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.250 |
0.050 |
| 175 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
-0.248 |
0.005 |
| 176 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
-0.246 |
0.032 |
| 177 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.246 |
-0.030 |
| 178 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
-0.245 |
-0.030 |
| 179 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.242 |
0.069 |
| 180 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
-0.241 |
-0.051 |
| 181 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
-0.239 |
0.025 |
| 182 |
249640_at |
AT5G36940
|
CAT3, cationic amino acid transporter 3 |
-0.237 |
0.021 |
| 183 |
247918_at |
AT5G57610
|
[Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain] |
-0.236 |
-0.013 |
| 184 |
259057_at |
AT3G03310
|
ATLCAT3, ARABIDOPSIS LECITHIN:CHOLESTEROL ACYLTRANSFERASE 3, LCAT3, lecithin:cholesterol acyltransferase 3 |
-0.236 |
0.009 |
| 185 |
258925_at |
AT3G10420
|
SPD1, SEEDLING PLASTID DEVELOPMENT 1 |
-0.236 |
-0.015 |
| 186 |
264341_at |
AT1G70270
|
unknown |
-0.234 |
-0.125 |
| 187 |
265898_at |
AT2G25690
|
unknown |
-0.234 |
-0.061 |