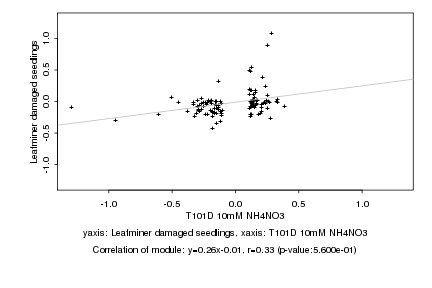
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
T101D 10mM NH4NO3 |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
0.386 |
-0.068 |
| 2 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
0.331 |
0.040 |
| 3 |
250576_at |
AT5G08250
|
[Cytochrome P450 superfamily protein] |
0.331 |
-0.001 |
| 4 |
254743_at |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
0.317 |
0.007 |
| 5 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.280 |
1.094 |
| 6 |
265151_at |
AT1G51340
|
[MATE efflux family protein] |
0.269 |
-0.266 |
| 7 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
0.268 |
-0.009 |
| 8 |
254090_at |
AT4G25010
|
AtSWEET14, SWEET14 |
0.258 |
0.002 |
| 9 |
250639_at |
AT5G07560
|
ATGRP20, GRP20, glycine-rich protein 20 |
0.253 |
0.098 |
| 10 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.249 |
0.894 |
| 11 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
0.247 |
-0.098 |
| 12 |
250605_at |
AT5G07570
|
[glycine/proline-rich protein] |
0.243 |
0.022 |
| 13 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.236 |
0.248 |
| 14 |
246374_at |
AT1G51840
|
[protein kinase-related] |
0.234 |
-0.021 |
| 15 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
0.229 |
-0.003 |
| 16 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.224 |
-0.012 |
| 17 |
262719_at |
AT1G43590
|
unknown |
0.218 |
-0.017 |
| 18 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.213 |
0.383 |
| 19 |
246617_at |
AT5G36270
|
[similar to DHAR2, glutathione dehydrogenase (ascorbate) [Arabidopsis thaliana] (TAIR:AT1G75270.1)] |
0.203 |
-0.090 |
| 20 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
0.200 |
-0.145 |
| 21 |
261541_at |
AT1G63600
|
[Receptor-like protein kinase-related family protein] |
0.198 |
-0.036 |
| 22 |
262898_at |
AT1G59850
|
[ARM repeat superfamily protein] |
0.191 |
-0.184 |
| 23 |
255957_at |
AT1G22160
|
unknown |
0.177 |
-0.196 |
| 24 |
258935_at |
AT3G10120
|
unknown |
0.167 |
0.019 |
| 25 |
260344_at |
AT1G69240
|
ATMES15, ARABIDOPSIS THALIANA METHYL ESTERASE 15, MES15, methyl esterase 15, RHS9, ROOT HAIR SPECIFIC 9 |
0.166 |
-0.045 |
| 26 |
261199_at |
AT1G12950
|
RSH2, root hair specific 2 |
0.161 |
-0.027 |
| 27 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
0.159 |
0.017 |
| 28 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
0.158 |
-0.051 |
| 29 |
258566_at |
AT3G04110
|
ATGLR1.1, GLR1, GLUTAMATE RECEPTOR 1, GLR1.1, glutamate receptor 1.1 |
0.154 |
0.175 |
| 30 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
0.152 |
0.153 |
| 31 |
256927_at |
AT3G22550
|
unknown |
0.149 |
-0.084 |
| 32 |
265246_at |
AT2G43050
|
ATPMEPCRD |
0.147 |
0.074 |
| 33 |
251689_at |
AT3G56500
|
[serine-rich protein-related] |
0.141 |
0.126 |
| 34 |
248569_at |
AT5G49770
|
[Leucine-rich repeat protein kinase family protein] |
0.140 |
0.003 |
| 35 |
252444_at |
AT3G46980
|
PHT4;3, phosphate transporter 4;3 |
0.136 |
-0.068 |
| 36 |
258620_at |
AT3G02940
|
AtMYB107, myb domain protein 107, MYB107, myb domain protein 107 |
0.135 |
0.045 |
| 37 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.132 |
-0.067 |
| 38 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
0.131 |
0.023 |
| 39 |
256563_at |
AT3G29780
|
RALFL27, ralf-like 27 |
0.128 |
-0.042 |
| 40 |
264054_at |
AT2G22540
|
AGL22, AGAMOUS-like 22, FAQ1, Flowering Arabidopsis QTL1, SVP, SHORT VEGETATIVE PHASE |
0.128 |
-0.029 |
| 41 |
260161_at |
AT1G79860
|
ATROPGEF12, MEE64, MATERNAL EFFECT EMBRYO ARREST 64, ROPGEF12, ROP (rho of plants) guanine nucleotide exchange factor 12 |
0.127 |
-0.014 |
| 42 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.124 |
0.552 |
| 43 |
259720_at |
AT1G61080
|
[Hydroxyproline-rich glycoprotein family protein] |
0.124 |
-0.001 |
| 44 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
0.123 |
-0.020 |
| 45 |
263817_at |
AT2G10070
|
unknown |
0.121 |
-0.066 |
| 46 |
249263_at |
AT5G41730
|
[Protein kinase family protein] |
0.120 |
0.183 |
| 47 |
267160_at |
AT2G37670
|
[Transducin/WD40 repeat-like superfamily protein] |
0.120 |
0.005 |
| 48 |
266004_at |
AT2G37330
|
ALS3, ALUMINUM SENSITIVE 3 |
0.120 |
-0.203 |
| 49 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
0.115 |
-0.234 |
| 50 |
267207_at |
AT2G30840
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.114 |
0.183 |
| 51 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
0.113 |
-0.070 |
| 52 |
250631_at |
AT5G07430
|
[Pectin lyase-like superfamily protein] |
0.112 |
0.009 |
| 53 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.112 |
0.491 |
| 54 |
264348_at |
AT1G12110
|
AtNPF6.3, ATNRT1, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 1, B-1, CHL1, CHLORINA 1, CHL1-1, NPF6.3, NRT1/ PTR family 6.3, NRT1, NITRATE TRANSPORTER 1, NRT1.1, nitrate transporter 1.1 |
0.111 |
-0.194 |
| 55 |
249791_at |
AT5G23810
|
AAP7, amino acid permease 7 |
0.109 |
0.193 |
| 56 |
265327_at |
AT2G18210
|
unknown |
0.109 |
0.117 |
| 57 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
0.108 |
-0.109 |
| 58 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.108 |
0.502 |
| 59 |
264371_at |
AT1G12090
|
ELP, extensin-like protein |
-1.300 |
-0.088 |
| 60 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.949 |
-0.287 |
| 61 |
262753_at |
AT1G16340
|
ATKDSA2, ATKSDA |
-0.613 |
-0.202 |
| 62 |
251319_at |
AT3G61610
|
[Galactose mutarotase-like superfamily protein] |
-0.511 |
0.079 |
| 63 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.456 |
-0.013 |
| 64 |
255720_at |
AT1G32060
|
PRK, phosphoribulokinase |
-0.382 |
-0.152 |
| 65 |
263114_at |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
-0.339 |
-0.036 |
| 66 |
254790_at |
AT4G12800
|
PSAL, photosystem I subunit l |
-0.337 |
-0.006 |
| 67 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
-0.326 |
-0.235 |
| 68 |
251762_at |
AT3G55800
|
SBPASE, sedoheptulose-bisphosphatase |
-0.309 |
-0.182 |
| 69 |
251229_at |
AT3G62740
|
BGLU7, beta glucosidase 7 |
-0.303 |
0.020 |
| 70 |
264837_at |
AT1G03600
|
PSB27 |
-0.300 |
-0.076 |
| 71 |
262970_at |
AT1G75690
|
LQY1, LOW QUANTUM YIELD OF PHOTOSYSTEM II 1 |
-0.294 |
-0.141 |
| 72 |
265998_at |
AT2G24270
|
ALDH11A3, aldehyde dehydrogenase 11A3 |
-0.292 |
-0.121 |
| 73 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
-0.291 |
-0.152 |
| 74 |
257807_at |
AT3G26650
|
GAPA, glyceraldehyde 3-phosphate dehydrogenase A subunit, GAPA-1, GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT 1 |
-0.287 |
-0.062 |
| 75 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
-0.274 |
0.052 |
| 76 |
258285_at |
AT3G16140
|
PSAH-1, photosystem I subunit H-1 |
-0.270 |
-0.030 |
| 77 |
259625_at |
AT1G42970
|
GAPB, glyceraldehyde-3-phosphate dehydrogenase B subunit |
-0.268 |
-0.119 |
| 78 |
255435_at |
AT4G03280
|
PETC, photosynthetic electron transfer C, PGR1, PROTON GRADIENT REGULATION 1 |
-0.258 |
-0.064 |
| 79 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
-0.253 |
-0.006 |
| 80 |
245701_at |
AT5G04140
|
FD-GOGAT, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE, GLS1, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1, GLU1, glutamate synthase 1, GLUS |
-0.242 |
-0.200 |
| 81 |
253738_at |
AT4G28750
|
PSAE-1, PSA E1 KNOCKOUT |
-0.241 |
-0.006 |
| 82 |
264185_at |
AT1G54780
|
AtTLP18.3, TLP18.3, thylakoid lumen protein 18.3 |
-0.232 |
-0.035 |
| 83 |
254970_at |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
-0.225 |
-0.003 |
| 84 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.221 |
-0.190 |
| 85 |
261746_at |
AT1G08380
|
PSAO, photosystem I subunit O |
-0.220 |
0.024 |
| 86 |
251814_at |
AT3G54890
|
LHCA1, photosystem I light harvesting complex gene 1 |
-0.200 |
0.001 |
| 87 |
263676_at |
AT1G09340
|
CRB, chloroplast RNA binding, CSP41B, CHLOROPLAST STEM-LOOP BINDING PROTEIN OF 41 KDA, HIP1.3, heteroglycan-interacting protein 1.3 |
-0.199 |
-0.133 |
| 88 |
245061_at |
AT2G39730
|
RCA, rubisco activase |
-0.195 |
-0.028 |
| 89 |
247649_at |
AT5G60030
|
unknown |
-0.195 |
0.025 |
| 90 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.187 |
-0.416 |
| 91 |
262283_at |
AT1G68590
|
PSRP3/1, plastid-specific ribosomal protein 3/1 |
-0.187 |
-0.173 |
| 92 |
266636_at |
AT2G35370
|
GDCH, glycine decarboxylase complex H |
-0.185 |
-0.158 |
| 93 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.184 |
-0.235 |
| 94 |
254102_at |
AT4G25050
|
ACP4, acyl carrier protein 4 |
-0.171 |
-0.097 |
| 95 |
260704_at |
AT1G32470
|
[Single hybrid motif superfamily protein] |
-0.169 |
-0.189 |
| 96 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
-0.167 |
-0.168 |
| 97 |
255886_at |
AT1G20340
|
DRT112, DNA-DAMAGE-REPAIR/TOLERATION PROTEIN 112, PETE2, PLASTOCYANIN 2 |
-0.164 |
-0.043 |
| 98 |
261218_at |
AT1G20020
|
ATLFNR2, LEAF FNR 2, FNR2, ferredoxin-NADP(+)-oxidoreductase 2, LFNR2, leaf-type chloroplast-targeted FNR 2 |
-0.158 |
0.007 |
| 99 |
265033_at |
AT1G61520
|
LHCA3, photosystem I light harvesting complex gene 3 |
-0.155 |
0.006 |
| 100 |
255078_at |
AT4G09010
|
APX4, ascorbate peroxidase 4, TL29, thylakoid lumen 29 |
-0.154 |
-0.104 |
| 101 |
252353_at |
AT3G48200
|
unknown |
-0.153 |
-0.348 |
| 102 |
251243_at |
AT3G61870
|
unknown |
-0.152 |
-0.184 |
| 103 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.150 |
-0.053 |
| 104 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
-0.140 |
0.324 |
| 105 |
267612_at |
AT2G26690
|
AtNPF6.2, NPF6.2, NRT1/ PTR family 6.2 |
-0.139 |
-0.111 |
| 106 |
248408_at |
AT5G51520
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.136 |
-0.060 |
| 107 |
259038_at |
AT3G09210
|
PTAC13, plastid transcriptionally active 13 |
-0.135 |
-0.087 |
| 108 |
266642_at |
AT2G35410
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.134 |
-0.122 |
| 109 |
265182_at |
AT1G23740
|
AOR, alkenal/one oxidoreductase |
-0.122 |
-0.158 |
| 110 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.120 |
-0.304 |
| 111 |
248201_at |
AT5G54180
|
PTAC15, plastid transcriptionally active 15 |
-0.119 |
0.008 |
| 112 |
251031_at |
AT5G02120
|
OHP, one helix protein, PDE335, PIGMENT DEFECTIVE 335 |
-0.116 |
-0.179 |
| 113 |
257773_at |
AT3G29185
|
unknown |
-0.115 |
-0.216 |
| 114 |
245023_at |
ATCG00080
|
PSBI, photosystem II reaction center protein I |
-0.114 |
-0.028 |
| 115 |
254137_at |
AT4G24930
|
[thylakoid lumenal 17.9 kDa protein, chloroplast] |
-0.110 |
-0.141 |