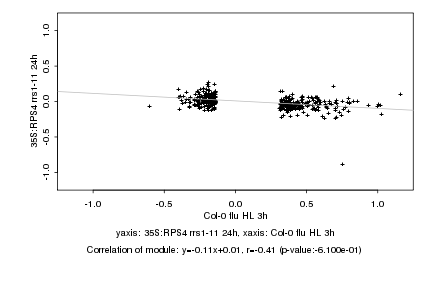
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 flu HL 3h |
Log2 signal ratio
35S:RPS4 rrs1-11 24h |
| 1 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
1.157 |
0.106 |
| 2 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.020 |
-0.172 |
| 3 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
1.013 |
-0.053 |
| 4 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
1.001 |
-0.034 |
| 5 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.998 |
-0.062 |
| 6 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.928 |
-0.043 |
| 7 |
255811_at |
AT4G10250
|
ATHSP22.0 |
0.857 |
0.007 |
| 8 |
252739_at |
AT3G43250
|
unknown |
0.829 |
0.007 |
| 9 |
267449_at |
AT2G33690
|
[Late embryogenesis abundant protein, group 6] |
0.801 |
-0.005 |
| 10 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
0.799 |
-0.020 |
| 11 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.794 |
-0.132 |
| 12 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
0.793 |
0.052 |
| 13 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.782 |
-0.009 |
| 14 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.768 |
-0.076 |
| 15 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.754 |
-0.104 |
| 16 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.750 |
-0.884 |
| 17 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.749 |
0.014 |
| 18 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.745 |
-0.184 |
| 19 |
251114_at |
AT5G01380
|
[Homeodomain-like superfamily protein] |
0.728 |
-0.149 |
| 20 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.712 |
-0.070 |
| 21 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.709 |
-0.025 |
| 22 |
263147_at |
AT1G53980
|
[Ubiquitin-like superfamily protein] |
0.709 |
0.021 |
| 23 |
246823_at |
AT5G26970
|
unknown |
0.705 |
-0.018 |
| 24 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.705 |
-0.224 |
| 25 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.703 |
-0.121 |
| 26 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
0.703 |
-0.230 |
| 27 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
0.702 |
-0.008 |
| 28 |
260744_at |
AT1G15010
|
unknown |
0.702 |
-0.095 |
| 29 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
0.682 |
0.217 |
| 30 |
256991_at |
AT3G28600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.670 |
-0.039 |
| 31 |
253689_at |
AT4G29770
|
unknown |
0.668 |
-0.036 |
| 32 |
252364_at |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.658 |
0.009 |
| 33 |
264718_at |
AT1G70130
|
LecRK-V.2, L-type lectin receptor kinase V.2 |
0.658 |
-0.018 |
| 34 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.647 |
-0.165 |
| 35 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.644 |
-0.085 |
| 36 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.637 |
-0.073 |
| 37 |
266486_at |
AT2G47950
|
unknown |
0.634 |
-0.066 |
| 38 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.629 |
-0.033 |
| 39 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.624 |
-0.239 |
| 40 |
256364_at |
AT1G66550
|
ATWRKY67, WRKY67, WRKY DNA-binding protein 67 |
0.621 |
0.002 |
| 41 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.608 |
-0.207 |
| 42 |
245363_at |
AT4G15120
|
[VQ motif-containing protein] |
0.595 |
-0.002 |
| 43 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.588 |
0.015 |
| 44 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.586 |
-0.029 |
| 45 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.583 |
-0.078 |
| 46 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.580 |
-0.119 |
| 47 |
261430_at |
AT1G18830
|
SEC31A |
0.577 |
0.035 |
| 48 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.576 |
-0.047 |
| 49 |
265482_at |
AT2G15780
|
[Cupredoxin superfamily protein] |
0.571 |
0.006 |
| 50 |
246537_at |
AT5G15940
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.570 |
0.060 |
| 51 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
0.569 |
0.060 |
| 52 |
245076_at |
AT2G23170
|
GH3.3 |
0.564 |
-0.107 |
| 53 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.561 |
-0.016 |
| 54 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.559 |
-0.009 |
| 55 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.559 |
-0.118 |
| 56 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.556 |
0.061 |
| 57 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.551 |
-0.109 |
| 58 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.549 |
-0.094 |
| 59 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.546 |
0.018 |
| 60 |
252882_at |
AT4G39675
|
unknown |
0.544 |
-0.086 |
| 61 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
0.538 |
-0.044 |
| 62 |
252036_at |
AT3G52070
|
unknown |
0.538 |
0.047 |
| 63 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.535 |
-0.101 |
| 64 |
264252_at |
AT1G09180
|
ATSAR1, SECRETION-ASSOCIATED RAS 1, ATSARA1A, secretion-associated RAS super family 1, SARA1A, secretion-associated RAS super family 1 |
0.533 |
0.062 |
| 65 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
0.520 |
0.007 |
| 66 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.513 |
-0.043 |
| 67 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.510 |
0.063 |
| 68 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.510 |
-0.038 |
| 69 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.509 |
-0.039 |
| 70 |
246406_at |
AT1G57650
|
[ATP binding] |
0.506 |
-0.187 |
| 71 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.505 |
-0.012 |
| 72 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.505 |
-0.031 |
| 73 |
257835_at |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
0.501 |
-0.066 |
| 74 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
0.496 |
-0.023 |
| 75 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.489 |
-0.024 |
| 76 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
0.486 |
-0.040 |
| 77 |
251761_at |
AT3G55700
|
[UDP-Glycosyltransferase superfamily protein] |
0.476 |
-0.154 |
| 78 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.470 |
0.036 |
| 79 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.469 |
-0.092 |
| 80 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.469 |
-0.012 |
| 81 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.468 |
-0.057 |
| 82 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.467 |
-0.033 |
| 83 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.465 |
0.057 |
| 84 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.465 |
0.077 |
| 85 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.465 |
-0.038 |
| 86 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.464 |
-0.080 |
| 87 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.464 |
0.063 |
| 88 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.463 |
-0.072 |
| 89 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
0.463 |
0.028 |
| 90 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
0.462 |
-0.083 |
| 91 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.451 |
-0.080 |
| 92 |
245243_at |
AT1G44414
|
unknown |
0.449 |
-0.042 |
| 93 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.448 |
-0.059 |
| 94 |
247509_at |
AT5G62020
|
AT-HSFB2A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR B2A, HSFB2A, heat shock transcription factor B2A |
0.448 |
-0.053 |
| 95 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
0.447 |
-0.091 |
| 96 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.447 |
-0.112 |
| 97 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.446 |
-0.047 |
| 98 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.446 |
-0.038 |
| 99 |
246418_at |
AT5G16960
|
[Zinc-binding dehydrogenase family protein] |
0.444 |
-0.084 |
| 100 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.442 |
-0.015 |
| 101 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.441 |
-0.000 |
| 102 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.438 |
-0.042 |
| 103 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.435 |
-0.196 |
| 104 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.433 |
-0.085 |
| 105 |
256045_at |
AT1G07150
|
MAPKKK13, mitogen-activated protein kinase kinase kinase 13 |
0.430 |
0.008 |
| 106 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.428 |
-0.056 |
| 107 |
263877_at |
AT2G21780
|
unknown |
0.428 |
-0.014 |
| 108 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.426 |
-0.043 |
| 109 |
266469_at |
AT2G31180
|
ATMYB14, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 14, MYB14, myb domain protein 14, MYB14AT |
0.424 |
-0.039 |
| 110 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.422 |
-0.030 |
| 111 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.422 |
-0.027 |
| 112 |
257994_at |
AT3G19920
|
unknown |
0.419 |
-0.051 |
| 113 |
264580_at |
AT1G05340
|
unknown |
0.419 |
-0.032 |
| 114 |
263742_at |
AT2G20625
|
unknown |
0.419 |
-0.026 |
| 115 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.417 |
-0.104 |
| 116 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.413 |
-0.063 |
| 117 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.413 |
-0.091 |
| 118 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.411 |
-0.068 |
| 119 |
262085_at |
AT1G56060
|
unknown |
0.411 |
-0.071 |
| 120 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.408 |
-0.074 |
| 121 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.405 |
-0.004 |
| 122 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.402 |
-0.063 |
| 123 |
246584_at |
AT5G14730
|
unknown |
0.398 |
0.112 |
| 124 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.397 |
-0.105 |
| 125 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.396 |
0.047 |
| 126 |
263772_at |
AT2G06255
|
ELF4-L3, ELF4-like 3 |
0.394 |
-0.062 |
| 127 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.393 |
-0.079 |
| 128 |
259947_at |
AT1G71530
|
[Protein kinase superfamily protein] |
0.392 |
-0.031 |
| 129 |
246018_at |
AT5G10695
|
unknown |
0.389 |
-0.068 |
| 130 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.387 |
0.040 |
| 131 |
261241_at |
AT1G32950
|
[Subtilase family protein] |
0.386 |
-0.123 |
| 132 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.384 |
-0.007 |
| 133 |
250211_at |
AT5G13880
|
unknown |
0.383 |
-0.209 |
| 134 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.382 |
-0.049 |
| 135 |
258337_at |
AT3G16040
|
[Translation machinery associated TMA7] |
0.382 |
-0.015 |
| 136 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.381 |
-0.043 |
| 137 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
0.380 |
-0.083 |
| 138 |
267493_at |
AT2G30400
|
ATOFP2, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 2, OFP2, ovate family protein 2 |
0.380 |
-0.032 |
| 139 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.380 |
-0.046 |
| 140 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.379 |
0.023 |
| 141 |
266017_at |
AT2G18690
|
unknown |
0.378 |
-0.033 |
| 142 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.377 |
-0.084 |
| 143 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.374 |
-0.057 |
| 144 |
254286_at |
AT4G22950
|
AGL19, AGAMOUS-like 19, GL19 |
0.374 |
0.027 |
| 145 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.373 |
0.081 |
| 146 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.372 |
0.056 |
| 147 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.371 |
-0.114 |
| 148 |
251422_at |
AT3G60540
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
0.369 |
0.019 |
| 149 |
259478_at |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
0.368 |
0.002 |
| 150 |
259032_at |
AT3G09380
|
unknown |
0.368 |
0.008 |
| 151 |
258610_at |
AT3G02875
|
ILR1, IAA-LEUCINE RESISTANT 1 |
0.367 |
-0.038 |
| 152 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
0.367 |
0.000 |
| 153 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
0.366 |
-0.038 |
| 154 |
247177_at |
AT5G65300
|
unknown |
0.366 |
-0.111 |
| 155 |
251207_at |
AT3G63050
|
unknown |
0.364 |
-0.039 |
| 156 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.362 |
-0.032 |
| 157 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.361 |
-0.008 |
| 158 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.361 |
-0.143 |
| 159 |
254857_at |
AT4G12120
|
ATSEC1B, SEC1B |
0.360 |
-0.082 |
| 160 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.360 |
-0.018 |
| 161 |
261069_at |
AT1G07410
|
ATRAB-A2B, ARABIDOPSIS RAB GTPASE HOMOLOG A2B, ATRABA2B, RAB GTPase homolog A2B, RAB-A2B, RAB GTPASE HOMOLOG A2B, RABA2b, RAB GTPase homolog A2B |
0.359 |
-0.019 |
| 162 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.358 |
0.063 |
| 163 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
0.357 |
-0.035 |
| 164 |
248959_at |
AT5G45630
|
unknown |
0.357 |
-0.118 |
| 165 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.356 |
-0.053 |
| 166 |
251156_at |
AT3G63060
|
EDL3, EID1-like 3 |
0.356 |
0.060 |
| 167 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.355 |
0.016 |
| 168 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.355 |
-0.016 |
| 169 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.353 |
-0.023 |
| 170 |
257874_at |
AT3G17110
|
[pseudogene] |
0.353 |
-0.068 |
| 171 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.353 |
0.003 |
| 172 |
260243_at |
AT1G63720
|
unknown |
0.352 |
-0.083 |
| 173 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.351 |
-0.033 |
| 174 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.351 |
-0.040 |
| 175 |
261236_at |
AT1G32880
|
[ARM repeat superfamily protein] |
0.346 |
0.058 |
| 176 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.345 |
-0.016 |
| 177 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.344 |
-0.111 |
| 178 |
258189_at |
AT3G17860
|
JAI3, JASMONATE-INSENSITIVE 3, JAZ3, jasmonate-zim-domain protein 3, TIFY6B |
0.342 |
-0.041 |
| 179 |
266975_at |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
0.342 |
0.022 |
| 180 |
260668_at |
AT1G19530
|
unknown |
0.340 |
-0.036 |
| 181 |
258939_at |
AT3G10020
|
unknown |
0.339 |
-0.010 |
| 182 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.337 |
-0.049 |
| 183 |
258262_at |
AT3G15770
|
unknown |
0.337 |
-0.060 |
| 184 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.337 |
-0.065 |
| 185 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
0.336 |
0.036 |
| 186 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.336 |
-0.069 |
| 187 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.331 |
-0.189 |
| 188 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.331 |
-0.048 |
| 189 |
254210_at |
AT4G23450
|
AIRP1, ABA Insensitive RING Protein 1, AtAIRP1 |
0.330 |
-0.042 |
| 190 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.328 |
0.145 |
| 191 |
246779_at |
AT5G27520
|
AtPNC2, PNC2, peroxisomal adenine nucleotide carrier 2 |
0.328 |
-0.089 |
| 192 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.327 |
-0.020 |
| 193 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.323 |
-0.039 |
| 194 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
0.323 |
0.037 |
| 195 |
264726_at |
AT1G22985
|
CRF7, cytokinin response factor 7 |
0.322 |
-0.055 |
| 196 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.322 |
-0.022 |
| 197 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.322 |
0.011 |
| 198 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.321 |
-0.085 |
| 199 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
0.320 |
-0.061 |
| 200 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.320 |
-0.024 |
| 201 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.318 |
0.028 |
| 202 |
258005_at |
AT3G19390
|
[Granulin repeat cysteine protease family protein] |
0.317 |
-0.223 |
| 203 |
260897_at |
AT1G29330
|
AERD2, ARABIDOPSIS ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ATERD2, ARABIDOPSIS THALIANA ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ERD2, ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2 |
0.315 |
-0.023 |
| 204 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
0.315 |
-0.113 |
| 205 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.314 |
-0.042 |
| 206 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.314 |
-0.071 |
| 207 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.312 |
-0.091 |
| 208 |
246370_at |
AT1G51920
|
unknown |
0.311 |
0.146 |
| 209 |
258760_at |
AT3G10780
|
[emp24/gp25L/p24 family/GOLD family protein] |
0.309 |
-0.090 |
| 210 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
-0.605 |
-0.060 |
| 211 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
-0.403 |
0.178 |
| 212 |
245672_at |
AT1G56710
|
[Pectin lyase-like superfamily protein] |
-0.403 |
0.060 |
| 213 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.398 |
0.060 |
| 214 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.397 |
0.070 |
| 215 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
-0.394 |
-0.102 |
| 216 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.390 |
0.081 |
| 217 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.381 |
0.016 |
| 218 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
-0.376 |
-0.024 |
| 219 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.366 |
0.075 |
| 220 |
249726_at |
AT5G35480
|
unknown |
-0.355 |
-0.005 |
| 221 |
245990_at |
AT5G20640
|
unknown |
-0.346 |
0.029 |
| 222 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.345 |
0.128 |
| 223 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.330 |
0.035 |
| 224 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.326 |
-0.062 |
| 225 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.325 |
-0.079 |
| 226 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.324 |
-0.004 |
| 227 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.317 |
-0.009 |
| 228 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.316 |
0.057 |
| 229 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.293 |
-0.051 |
| 230 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.292 |
-0.042 |
| 231 |
247212_at |
AT5G65040
|
unknown |
-0.288 |
0.038 |
| 232 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.288 |
0.018 |
| 233 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.285 |
-0.051 |
| 234 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.282 |
0.102 |
| 235 |
252236_at |
AT3G49930
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.274 |
0.018 |
| 236 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.272 |
-0.036 |
| 237 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.270 |
0.142 |
| 238 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.270 |
0.012 |
| 239 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.268 |
0.061 |
| 240 |
256099_at |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
-0.267 |
-0.054 |
| 241 |
247225_at |
AT5G65090
|
BST1, BRISTLED 1, DER4, DEFORMED ROOT HAIRS 4, MRH3 |
-0.266 |
-0.065 |
| 242 |
250102_at |
AT5G16590
|
LRR1, Leucine rich repeat protein 1 |
-0.265 |
0.128 |
| 243 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
-0.265 |
-0.008 |
| 244 |
249420_at |
AT5G39820
|
anac094, NAC domain containing protein 94, NAC094, NAC domain containing protein 94 |
-0.264 |
-0.059 |
| 245 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.262 |
0.134 |
| 246 |
251292_at |
AT3G61920
|
unknown |
-0.262 |
0.003 |
| 247 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.258 |
0.108 |
| 248 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.256 |
0.042 |
| 249 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.256 |
-0.042 |
| 250 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.254 |
-0.015 |
| 251 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.253 |
-0.095 |
| 252 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.253 |
0.079 |
| 253 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.250 |
0.178 |
| 254 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.246 |
0.063 |
| 255 |
245690_at |
AT5G04230
|
ATPAL3, PAL3, phenyl alanine ammonia-lyase 3 |
-0.245 |
-0.014 |
| 256 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.239 |
0.083 |
| 257 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
-0.239 |
-0.080 |
| 258 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.237 |
0.015 |
| 259 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.235 |
-0.001 |
| 260 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.234 |
0.063 |
| 261 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.231 |
0.034 |
| 262 |
248443_at |
AT5G51310
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.231 |
0.003 |
| 263 |
263421_at |
AT2G17230
|
EXL5, EXORDIUM like 5 |
-0.230 |
0.033 |
| 264 |
257761_at |
AT3G23090
|
WDL3 |
-0.229 |
0.014 |
| 265 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.229 |
0.149 |
| 266 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.228 |
0.186 |
| 267 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
-0.225 |
-0.024 |
| 268 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
-0.223 |
0.098 |
| 269 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.223 |
-0.058 |
| 270 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.223 |
-0.113 |
| 271 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.223 |
0.044 |
| 272 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-0.222 |
0.063 |
| 273 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.221 |
0.091 |
| 274 |
251931_at |
AT3G53950
|
[glyoxal oxidase-related protein] |
-0.218 |
0.014 |
| 275 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.217 |
0.069 |
| 276 |
255443_at |
AT4G02700
|
SULTR3;2, sulfate transporter 3;2 |
-0.217 |
-0.004 |
| 277 |
249190_at |
AT5G42750
|
BKI1, BRI1 kinase inhibitor 1 |
-0.216 |
0.068 |
| 278 |
263675_x_at |
AT2G04770
|
[CACTA-like transposase family (Ptta/En/Spm), has a 6.0e-20 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.215 |
0.029 |
| 279 |
261596_at |
AT1G33080
|
[MATE efflux family protein] |
-0.213 |
0.026 |
| 280 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.212 |
0.124 |
| 281 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.210 |
0.084 |
| 282 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.209 |
0.181 |
| 283 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.208 |
0.036 |
| 284 |
245032_at |
AT2G26630
|
[transposase IS4 family protein, contains Pfam profile: PF01609 transposase DDE domain] |
-0.207 |
-0.023 |
| 285 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.205 |
0.116 |
| 286 |
255506_at |
AT4G02130
|
GATL6, galacturonosyltransferase-like 6, LGT10 |
-0.205 |
0.017 |
| 287 |
259574_at |
AT1G35310
|
MLP168, MLP-like protein 168 |
-0.204 |
-0.045 |
| 288 |
245029_at |
AT2G26580
|
YAB5, YABBY5 |
-0.204 |
0.122 |
| 289 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.202 |
-0.006 |
| 290 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.202 |
-0.093 |
| 291 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.200 |
0.246 |
| 292 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.200 |
-0.069 |
| 293 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.198 |
0.090 |
| 294 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.196 |
0.224 |
| 295 |
254274_at |
AT4G22770
|
AHL2, AT-hook motif nuclear localized protein 2 |
-0.196 |
-0.114 |
| 296 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
-0.196 |
0.162 |
| 297 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.195 |
0.100 |
| 298 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.195 |
0.281 |
| 299 |
264770_at |
AT1G23030
|
[ARM repeat superfamily protein] |
-0.194 |
0.012 |
| 300 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
-0.193 |
-0.052 |
| 301 |
267146_at |
AT2G38160
|
unknown |
-0.193 |
-0.026 |
| 302 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-0.192 |
0.059 |
| 303 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.192 |
0.033 |
| 304 |
263674_at |
AT2G04790
|
unknown |
-0.192 |
0.119 |
| 305 |
259566_at |
AT1G20520
|
unknown |
-0.191 |
0.018 |
| 306 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
-0.189 |
-0.065 |
| 307 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.189 |
0.074 |
| 308 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.188 |
0.167 |
| 309 |
250975_at |
AT5G03050
|
unknown |
-0.188 |
0.004 |
| 310 |
258596_at |
AT3G04510
|
LSH2, LIGHT SENSITIVE HYPOCOTYLS 2 |
-0.185 |
0.101 |
| 311 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.184 |
0.037 |
| 312 |
251995_at |
AT3G52940
|
ELL1, EXTRA-LONG-LIFESPAN 1, FK, FACKEL, HYD2 |
-0.183 |
0.074 |
| 313 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.183 |
-0.004 |
| 314 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.183 |
-0.077 |
| 315 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.182 |
0.021 |
| 316 |
247291_at |
AT5G64480
|
unknown |
-0.181 |
0.008 |
| 317 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.181 |
0.010 |
| 318 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
-0.180 |
0.041 |
| 319 |
247347_at |
AT5G63780
|
SHA1, shoot apical meristem arrest 1 |
-0.179 |
0.072 |
| 320 |
259815_at |
AT1G49870
|
unknown |
-0.178 |
-0.065 |
| 321 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.177 |
-0.000 |
| 322 |
251142_at |
AT5G01015
|
unknown |
-0.177 |
-0.008 |
| 323 |
257912_at |
AT3G25500
|
AFH1, formin homology 1, AHF1, ATFH1, ARABIDOPSIS THALIANA FORMIN HOMOLOGY 1, FH1, FORMIN HOMOLOGY 1 |
-0.176 |
0.094 |
| 324 |
262162_at |
AT1G78020
|
unknown |
-0.175 |
0.045 |
| 325 |
262704_at |
AT1G16530
|
ASL9, ASYMMETRIC LEAVES 2-like 9, LBD3, LOB DOMAIN-CONTAINING PROTEIN 3 |
-0.175 |
-0.115 |
| 326 |
245650_at |
AT1G24735
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.174 |
-0.030 |
| 327 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.173 |
0.019 |
| 328 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.173 |
0.078 |
| 329 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.172 |
0.116 |
| 330 |
265279_at |
AT2G28460
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.172 |
-0.004 |
| 331 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.172 |
-0.026 |
| 332 |
253050_at |
AT4G37450
|
AGP18, arabinogalactan protein 18, ATAGP18 |
-0.171 |
-0.054 |
| 333 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.171 |
0.121 |
| 334 |
248976_at |
AT5G44930
|
ARAD2, ARABINAN DEFICIENT 2 |
-0.171 |
0.018 |
| 335 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.170 |
0.126 |
| 336 |
249167_at |
AT5G42860
|
unknown |
-0.169 |
0.084 |
| 337 |
255578_at |
AT4G01450
|
UMAMIT30, Usually multiple acids move in and out Transporters 30 |
-0.169 |
0.064 |
| 338 |
251398_at |
AT3G60580
|
[C2H2-like zinc finger protein] |
-0.169 |
-0.027 |
| 339 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.169 |
0.006 |
| 340 |
254856_at |
AT4G12160
|
[pseudogene] |
-0.169 |
0.032 |
| 341 |
258435_at |
AT3G16740
|
[F-box and associated interaction domains-containing protein] |
-0.169 |
-0.032 |
| 342 |
262170_at |
AT1G74940
|
unknown |
-0.168 |
0.045 |
| 343 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.168 |
0.106 |
| 344 |
245185_at |
AT1G67760
|
[TCP-1/cpn60 chaperonin family protein] |
-0.168 |
-0.042 |
| 345 |
258633_at |
AT3G07990
|
SCPL27, serine carboxypeptidase-like 27 |
-0.167 |
0.047 |
| 346 |
263199_at |
AT1G05590
|
ATHEX3, BETA-HEXOSAMINIDASE 3, HEXO2, beta-hexosaminidase 2 |
-0.167 |
0.038 |
| 347 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.166 |
0.009 |
| 348 |
267535_at |
AT2G41940
|
ZFP8, zinc finger protein 8 |
-0.166 |
0.091 |
| 349 |
261075_at |
AT1G07280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.166 |
0.109 |
| 350 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.165 |
0.086 |
| 351 |
251646_at |
AT3G57780
|
unknown |
-0.165 |
0.039 |
| 352 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.165 |
0.037 |
| 353 |
262689_at |
AT1G62700
|
ANAC026, Arabidopsis NAC domain containing protein 26, VND5, VASCULAR RELATED NAC-DOMAIN PROTEIN 5 |
-0.164 |
0.005 |
| 354 |
248600_at |
AT5G49390
|
[LOCATED IN: endomembrane system] |
-0.164 |
-0.108 |
| 355 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.164 |
-0.030 |
| 356 |
253253_at |
AT4G34750
|
SAUR49, SMALL AUXIN UPREGULATED RNA 49 |
-0.164 |
0.036 |
| 357 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.163 |
0.011 |
| 358 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.163 |
0.023 |
| 359 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.163 |
0.018 |
| 360 |
252934_at |
AT4G39120
|
HISN7, HISTIDINE BIOSYNTHESIS 7, IMPL2, myo-inositol monophosphatase like 2 |
-0.162 |
0.065 |
| 361 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.162 |
0.003 |
| 362 |
250479_at |
AT5G10260
|
AtRABH1e, RAB GTPase homolog H1E, RABH1e, RAB GTPase homolog H1E |
-0.162 |
-0.044 |
| 363 |
254842_at |
AT4G11950
|
unknown |
-0.162 |
-0.041 |
| 364 |
249186_at |
AT5G43040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.161 |
-0.003 |
| 365 |
252707_at |
AT3G43790
|
ZIFL2, zinc induced facilitator-like 2 |
-0.161 |
-0.021 |
| 366 |
252712_at |
AT3G43800
|
ATGSTU27, glutathione S-transferase tau 27, GSTU27, glutathione S-transferase tau 27 |
-0.160 |
0.052 |
| 367 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.160 |
0.096 |
| 368 |
263915_at |
AT2G36430
|
unknown |
-0.160 |
0.057 |
| 369 |
252763_at |
AT3G42725
|
[Putative membrane lipoprotein] |
-0.158 |
0.066 |
| 370 |
248212_at |
AT5G54020
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.158 |
0.002 |
| 371 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
-0.157 |
-0.004 |
| 372 |
259160_at |
AT3G05410
|
[Photosystem II reaction center PsbP family protein] |
-0.157 |
0.091 |
| 373 |
260167_at |
AT1G71970
|
unknown |
-0.157 |
0.016 |
| 374 |
249235_at |
AT5G42100
|
ATBG_PPAP, ARABIDOPSIS THALIANA BETA-1,3-GLUCANASE_PUTATIVE, BG_PPAP, beta-1,3-glucanase_putative |
-0.157 |
0.049 |
| 375 |
249785_at |
AT5G24300
|
ATSS1, STARCH SYNTHASE 1, SS1, starch synthase 1 |
-0.156 |
0.045 |
| 376 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.155 |
0.040 |
| 377 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
-0.155 |
0.077 |
| 378 |
266889_at |
AT2G44640
|
unknown |
-0.155 |
0.013 |
| 379 |
245171_at |
AT2G47560
|
[RING/U-box superfamily protein] |
-0.154 |
-0.085 |
| 380 |
258782_at |
AT3G11750
|
FOLB1 |
-0.154 |
0.036 |
| 381 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.153 |
-0.004 |
| 382 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.153 |
-0.101 |
| 383 |
245417_at |
AT4G17360
|
[Formyl transferase] |
-0.153 |
0.022 |
| 384 |
261638_at |
AT1G49975
|
[INVOLVED IN: photosynthesis] |
-0.153 |
0.059 |
| 385 |
254930_at |
AT4G11450
|
unknown |
-0.152 |
-0.076 |
| 386 |
254810_at |
AT4G12390
|
PME1, pectin methylesterase inhibitor 1 |
-0.152 |
0.136 |
| 387 |
255751_at |
AT1G31950
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.152 |
0.020 |
| 388 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.152 |
0.058 |
| 389 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
-0.152 |
-0.031 |
| 390 |
248329_at |
AT5G52780
|
unknown |
-0.151 |
0.081 |
| 391 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.151 |
0.114 |
| 392 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
-0.150 |
0.113 |
| 393 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.150 |
0.103 |
| 394 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.150 |
-0.011 |
| 395 |
250145_at |
AT5G14690
|
unknown |
-0.150 |
0.006 |
| 396 |
258027_at |
AT3G19515
|
unknown |
-0.150 |
0.015 |
| 397 |
266118_at |
AT2G02130
|
LCR68, low-molecular-weight cysteine-rich 68, PDF2.3 |
-0.150 |
0.100 |
| 398 |
258369_at |
AT3G14310
|
ATPME3, pectin methylesterase 3, OZS2, OVERLY ZINC SENSITIVE 2, PME3, pectin methylesterase 3 |
-0.149 |
0.070 |
| 399 |
258806_at |
AT3G04020
|
unknown |
-0.148 |
-0.038 |
| 400 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.148 |
-0.020 |
| 401 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.148 |
0.243 |
| 402 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.148 |
0.105 |
| 403 |
256919_at |
AT3G18970
|
MEF20, mitochondrial editing factor 20 |
-0.148 |
0.019 |
| 404 |
252786_at |
AT3G42670
|
CHR38, chromatin remodeling 38, CLSY, CLASSY1, CLSY1, CLASSY 1 |
-0.147 |
-0.009 |
| 405 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.147 |
-0.013 |
| 406 |
246475_at |
AT5G16720
|
MyoB3, myosin binding protein 3 |
-0.147 |
0.004 |
| 407 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
-0.147 |
-0.005 |
| 408 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
-0.146 |
0.152 |
| 409 |
248691_at |
AT5G48310
|
unknown |
-0.146 |
-0.022 |
| 410 |
249288_at |
AT5G41050
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.145 |
0.071 |
| 411 |
251261_at |
AT3G62110
|
[Pectin lyase-like superfamily protein] |
-0.145 |
0.008 |
| 412 |
263664_at |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
-0.145 |
0.152 |
| 413 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.145 |
0.024 |
| 414 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.145 |
0.059 |
| 415 |
250647_at |
AT5G06770
|
[KH domain-containing protein / zinc finger (CCCH type) family protein] |
-0.145 |
0.013 |
| 416 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.145 |
0.101 |
| 417 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
-0.145 |
0.135 |