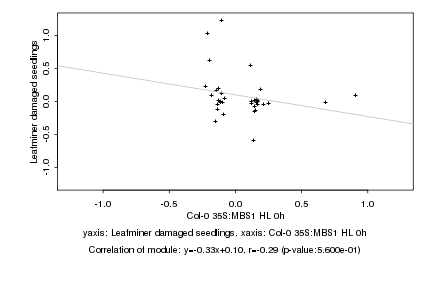
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 35S:MBS1 HL 0h |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
252036_at |
AT3G52070
|
unknown |
0.900 |
0.095 |
| 2 |
258623_at |
AT3G02790
|
MBS1, METHYLENE BLUE SENSITIVITY 1 |
0.677 |
-0.007 |
| 3 |
262363_at |
AT1G72850
|
[Disease resistance protein (TIR-NBS class)] |
0.245 |
-0.028 |
| 4 |
252755_at |
AT3G43530
|
unknown |
0.208 |
-0.037 |
| 5 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.185 |
0.185 |
| 6 |
255246_at |
AT4G05640
|
unknown |
0.164 |
-0.037 |
| 7 |
263725_at |
AT2G13550
|
unknown |
0.163 |
0.018 |
| 8 |
253622_at |
AT4G30560
|
ATCNGC9, cyclic nucleotide gated channel 9, CNGC9, cyclic nucleotide gated channel 9 |
0.161 |
0.006 |
| 9 |
260067_at |
AT1G73780
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.157 |
0.040 |
| 10 |
264739_at |
AT1G62260
|
MEF9, mitochondrial editing factor 9 |
0.155 |
-0.012 |
| 11 |
250393_at |
AT5G10900
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
0.150 |
-0.126 |
| 12 |
265554_at |
AT2G07505
|
[zinc ion binding] |
0.143 |
0.019 |
| 13 |
263718_at |
AT2G13570
|
NF-YB7, nuclear factor Y, subunit B7 |
0.143 |
-0.140 |
| 14 |
247701_at |
AT5G59900
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.137 |
-0.067 |
| 15 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
0.131 |
-0.588 |
| 16 |
245232_at |
AT4G25590
|
ADF7, actin depolymerizing factor 7 |
0.120 |
0.013 |
| 17 |
261732_at |
AT1G47770
|
[Beta-galactosidase related protein] |
0.114 |
-0.028 |
| 18 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.111 |
0.559 |
| 19 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.231 |
0.229 |
| 20 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.218 |
1.035 |
| 21 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
-0.199 |
0.626 |
| 22 |
247554_at |
AT5G61010
|
ATEXO70E2, exocyst subunit exo70 family protein E2, EXO70E2, exocyst subunit exo70 family protein E2 |
-0.184 |
0.096 |
| 23 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
-0.151 |
-0.303 |
| 24 |
245309_at |
AT4G15140
|
unknown |
-0.146 |
0.169 |
| 25 |
247714_at |
AT5G59340
|
WOX2, WUSCHEL related homeobox 2 |
-0.142 |
-0.116 |
| 26 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
-0.136 |
-0.042 |
| 27 |
262378_at |
AT1G72830
|
ATHAP2C, HAP2C, NF-YA3, nuclear factor Y, subunit A3 |
-0.134 |
0.201 |
| 28 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
-0.130 |
0.029 |
| 29 |
259828_at |
AT1G72220
|
[RING/U-box superfamily protein] |
-0.117 |
0.004 |
| 30 |
245128_at |
AT2G45380
|
unknown |
-0.114 |
-0.007 |
| 31 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.110 |
1.244 |
| 32 |
253807_at |
AT4G28280
|
LLG3, LORELEI-LIKE-GPI ANCHORED PROTEIN 3 |
-0.109 |
0.126 |
| 33 |
262831_at |
AT1G14730
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.103 |
-0.013 |
| 34 |
258587_at |
AT3G04310
|
unknown |
-0.094 |
-0.183 |
| 35 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
-0.090 |
0.046 |