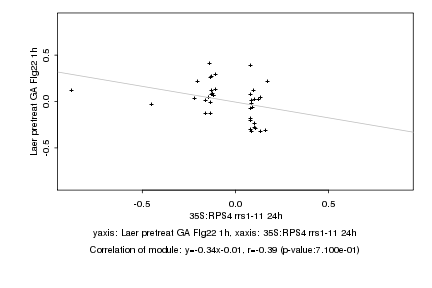
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S:RPS4 rrs1-11 24h |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.168 |
0.219 |
| 2 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
0.160 |
-0.311 |
| 3 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
0.134 |
-0.319 |
| 4 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
0.134 |
0.045 |
| 5 |
258828_at |
AT3G07130
|
ATPAP15, PURPLE ACID PHOSPHATASE 15, PAP15, purple acid phosphatase 15 |
0.123 |
0.024 |
| 6 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
0.106 |
-0.283 |
| 7 |
255876_at |
AT2G40480
|
unknown |
0.102 |
0.025 |
| 8 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
0.102 |
-0.235 |
| 9 |
258460_at |
AT3G17330
|
ECT6, evolutionarily conserved C-terminal region 6 |
0.097 |
-0.274 |
| 10 |
257764_at |
AT3G23010
|
AtRLP36, receptor like protein 36, RLP36, receptor like protein 36 |
0.096 |
0.124 |
| 11 |
267209_at |
AT2G30930
|
unknown |
0.091 |
-0.061 |
| 12 |
245070_at |
AT2G23240
|
AtMT4b, Arabidopsis thaliana metallothionein 4b |
0.086 |
-0.018 |
| 13 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
0.083 |
0.020 |
| 14 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
0.081 |
-0.317 |
| 15 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
0.079 |
-0.296 |
| 16 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
0.078 |
-0.201 |
| 17 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.078 |
-0.180 |
| 18 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.076 |
0.391 |
| 19 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
0.076 |
-0.065 |
| 20 |
253128_at |
AT4G36070
|
CPK18, calcium-dependent protein kinase 18 |
0.076 |
0.079 |
| 21 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.884 |
0.124 |
| 22 |
263658_at |
AT1G04490
|
unknown |
-0.455 |
-0.024 |
| 23 |
259923_at |
AT1G72760
|
[Protein kinase superfamily protein] |
-0.221 |
0.036 |
| 24 |
250211_at |
AT5G13880
|
unknown |
-0.209 |
0.221 |
| 25 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
-0.163 |
-0.123 |
| 26 |
250722_at |
AT5G06190
|
unknown |
-0.162 |
0.012 |
| 27 |
261313_at |
AT1G52970
|
DD11, downregulated in DIF1 11 |
-0.145 |
0.053 |
| 28 |
252448_at |
AT3G47050
|
[Glycosyl hydrolase family protein] |
-0.143 |
0.410 |
| 29 |
245801_at |
AT1G46912
|
[F-box associated ubiquitination effector family protein] |
-0.136 |
-0.010 |
| 30 |
247049_at |
AT5G66440
|
unknown |
-0.135 |
-0.123 |
| 31 |
252881_at |
AT4G39610
|
unknown |
-0.134 |
0.259 |
| 32 |
259370_at |
AT1G69050
|
unknown |
-0.130 |
0.272 |
| 33 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.129 |
0.083 |
| 34 |
254009_at |
AT4G26390
|
[Pyruvate kinase family protein] |
-0.129 |
0.128 |
| 35 |
262789_at |
AT1G10810
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.125 |
0.092 |
| 36 |
252571_at |
AT3G45280
|
ATSYP72, SYP72, syntaxin of plants 72 |
-0.119 |
0.073 |
| 37 |
257274_at |
AT3G14510
|
[Polyprenyl synthetase family protein] |
-0.118 |
0.067 |
| 38 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
-0.111 |
0.130 |
| 39 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
-0.110 |
0.302 |